In vivo protein trapping produces a functional expression codex of the vertebrate proteome
- PMID: 21552255
- PMCID: PMC3306164
- DOI: 10.1038/nmeth.1606
In vivo protein trapping produces a functional expression codex of the vertebrate proteome
Abstract
We describe a conditional in vivo protein-trap mutagenesis system that reveals spatiotemporal protein expression dynamics and can be used to assess gene function in the vertebrate Danio rerio. Integration of pGBT-RP2.1 (RP2), a gene-breaking transposon containing a protein trap, efficiently disrupts gene expression with >97% knockdown of normal transcript amounts and simultaneously reports protein expression for each locus. The mutant alleles are revertible in somatic tissues via Cre recombinase or splice-site-blocking morpholinos and are thus to our knowledge the first systematic conditional mutant alleles outside the mouse model. We report a collection of 350 zebrafish lines that include diverse molecular loci. RP2 integrations reveal the complexity of genomic architecture and gene function in a living organism and can provide information on protein subcellular localization. The RP2 mutagenesis system is a step toward a unified 'codex' of protein expression and direct functional annotation of the vertebrate genome.
Figures
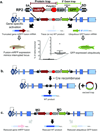
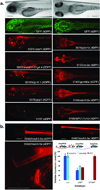
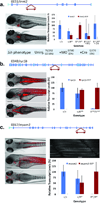
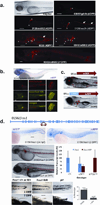
Comment in
-
All-in-one live: genes trapped, tagged and conditionally broken.Nat Methods. 2011 Jun;8(6):466-7. doi: 10.1038/nmeth.1617. Nat Methods. 2011. PMID: 21623352 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

