Genome partitioning of genetic variation for complex traits using common SNPs
- PMID: 21552263
- PMCID: PMC4295936
- DOI: 10.1038/ng.823
Genome partitioning of genetic variation for complex traits using common SNPs
Abstract
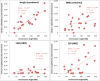
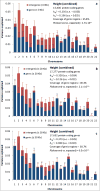
We estimate and partition genetic variation for height, body mass index (BMI), von Willebrand factor and QT interval (QTi) using 586,898 SNPs genotyped on 11,586 unrelated individuals. We estimate that ∼45%, ∼17%, ∼25% and ∼21% of the variance in height, BMI, von Willebrand factor and QTi, respectively, can be explained by all autosomal SNPs and a further ∼0.5-1% can be explained by X chromosome SNPs. We show that the variance explained by each chromosome is proportional to its length, and that SNPs in or near genes explain more variation than SNPs between genes. We propose a new approach to estimate variation due to cryptic relatedness and population stratification. Our results provide further evidence that a substantial proportion of heritability is captured by common SNPs, that height, BMI and QTi are highly polygenic traits, and that the additive variation explained by a part of the genome is approximately proportional to the total length of DNA contained within genes therein.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

