Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants
- PMID: 21552272
- PMCID: PMC3270583
- DOI: 10.1038/nbt.1852
Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants
Abstract
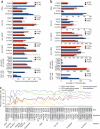
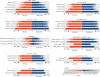
We have systematically compared copy number variant (CNV) detection on eleven microarrays to evaluate data quality and CNV calling, reproducibility, concordance across array platforms and laboratory sites, breakpoint accuracy and analysis tool variability. Different analytic tools applied to the same raw data typically yield CNV calls with <50% concordance. Moreover, reproducibility in replicate experiments is <70% for most platforms. Nevertheless, these findings should not preclude detection of large CNVs for clinical diagnostic purposes because large CNVs with poor reproducibility are found primarily in complex genomic regions and would typically be removed by standard clinical data curation. The striking differences between CNV calls from different platforms and analytic tools highlight the importance of careful assessment of experimental design in discovery and association studies and of strict data curation and filtering in diagnostics. The CNV resource presented here allows independent data evaluation and provides a means to benchmark new algorithms.
Figures


References
-
- Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. - PubMed
-
- Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- Tuzun E, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. - PubMed
-
- Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet. Genome Res. 2006;115:205–214. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
- 213997/CAPMC/ CIHR/Canada
- 077008/WT_/Wellcome Trust/United Kingdom
- T32 HD007396/HD/NICHD NIH HHS/United States
- R01 HD055150/HD/NICHD NIH HHS/United States
- HG004221/HG/NHGRI NIH HHS/United States
- HD055150/HD/NICHD NIH HHS/United States
- 077014/WT_/Wellcome Trust/United Kingdom
- P01 HD068250/HD/NICHD NIH HHS/United States
- HD007396/HD/NICHD NIH HHS/United States
- U01 HG005209/HG/NHGRI NIH HHS/United States
- CA111560/CA/NCI NIH HHS/United States
- HG005209/HG/NHGRI NIH HHS/United States
- R01 CA111560/CA/NCI NIH HHS/United States
- P41 HG004221/HG/NHGRI NIH HHS/United States
- T32 GM007748/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

