Evolution and classification of the CRISPR-Cas systems
- PMID: 21552286
- PMCID: PMC3380444
- DOI: 10.1038/nrmicro2577
Evolution and classification of the CRISPR-Cas systems
Abstract
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated proteins) modules are adaptive immunity systems that are present in many archaea and bacteria. These defence systems are encoded by operons that have an extraordinarily diverse architecture and a high rate of evolution for both the cas genes and the unique spacer content. Here, we provide an updated analysis of the evolutionary relationships between CRISPR-Cas systems and Cas proteins. Three major types of CRISPR-Cas system are delineated, with a further division into several subtypes and a few chimeric variants. Given the complexity of the genomic architectures and the extremely dynamic evolution of the CRISPR-Cas systems, a unified classification of these systems should be based on multiple criteria. Accordingly, we propose a 'polythetic' classification that integrates the phylogenies of the most common cas genes, the sequence and organization of the CRISPR repeats and the architecture of the CRISPR-cas loci.
Figures
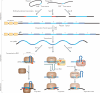
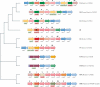
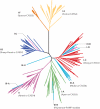
References
-
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 2010;64:475–493. - PubMed
-
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. - PubMed
-
- Sorek R, Kunin V, Hugenholtz P. CRISPR — a widespread system that provides acquired resistance against phages in bacteria and archaea. Nature Rev. Microbiol. 2008;6:181–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

