Deficiency of huntingtin has pleiotropic effects in the social amoeba Dictyostelium discoideum
- PMID: 21552328
- PMCID: PMC3084204
- DOI: 10.1371/journal.pgen.1002052
Deficiency of huntingtin has pleiotropic effects in the social amoeba Dictyostelium discoideum
Abstract
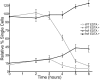
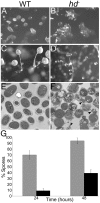
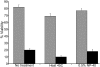
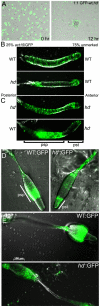
Huntingtin is a large HEAT repeat protein first identified in humans, where a polyglutamine tract expansion near the amino terminus causes a gain-of-function mechanism that leads to selective neuronal loss in Huntington's disease (HD). Genetic evidence in humans and knock-in mouse models suggests that this gain-of-function involves an increase or deregulation of some aspect of huntingtin's normal function(s), which remains poorly understood. As huntingtin shows evolutionary conservation, a powerful approach to discovering its normal biochemical role(s) is to study the effects caused by its deficiency in a model organism with a short life-cycle that comprises both cellular and multicellular developmental stages. To facilitate studies aimed at detailed knowledge of huntingtin's normal function(s), we generated a null mutant of hd, the HD ortholog in Dictyostelium discoideum. Dictyostelium cells lacking endogenous huntingtin were viable but during development did not exhibit the typical polarized morphology of Dictyostelium cells, streamed poorly to form aggregates by accretion rather than chemotaxis, showed disorganized F-actin staining, exhibited extreme sensitivity to hypoosmotic stress, and failed to form EDTA-resistant cell-cell contacts. Surprisingly, chemotactic streaming could be rescued in the presence of the bivalent cations Ca(2+) or Mg(2+) but not pulses of cAMP. Although hd(-) cells completed development, it was delayed and proceeded asynchronously, producing small fruiting bodies with round, defective spores that germinated spontaneously within a glassy sorus. When developed as chimeras with wild-type cells, hd(-) cells failed to populate the pre-spore region of the slug. In Dictyostelium, huntingtin deficiency is compatible with survival of the organism but renders cells sensitive to low osmolarity, which produces pleiotropic cell autonomous defects that affect cAMP signaling and as a consequence development. Thus, Dictyostelium provides a novel haploid organism model for genetic, cell biological, and biochemical studies to delineate the functions of the HD protein.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Huntingtin Supplies a csaA-Independent Function Essential for EDTA-Resistant Homotypic Cell Adhesion in Dictyostelium discoideum.J Huntingtons Dis. 2014;3(3):261-71. doi: 10.3233/JHD-140112. J Huntingtons Dis. 2014. PMID: 25300330
-
ForC lacks canonical formin activity but bundles actin filaments and is required for multicellular development of Dictyostelium cells.Eur J Cell Biol. 2013 Jun-Jul;92(6-7):201-12. doi: 10.1016/j.ejcb.2013.07.001. Epub 2013 Jul 11. Eur J Cell Biol. 2013. PMID: 23906540
-
Huntingtin regulates Ca(2+) chemotaxis and K(+)-facilitated cAMP chemotaxis, in conjunction with the monovalent cation/H(+) exchanger Nhe1, in a model developmental system: insights into its possible role in Huntington׳s disease.Dev Biol. 2014 Oct 1;394(1):24-38. doi: 10.1016/j.ydbio.2014.08.009. Epub 2014 Aug 19. Dev Biol. 2014. PMID: 25149514
-
Comparative analysis of spore coat formation, structure, and function in Dictyostelium.Int Rev Cytol. 2003;222:237-93. doi: 10.1016/s0074-7696(02)22016-1. Int Rev Cytol. 2003. PMID: 12503851 Review.
-
Proteomics opens doors to the mechanisms of developmentally regulated secretion.Mol Cell Proteomics. 2003 Nov;2(11):1156-63. doi: 10.1074/mcp.R300011-MCP200. Epub 2003 Sep 22. Mol Cell Proteomics. 2003. PMID: 14504294 Review.
Cited by
-
Using the social amoeba Dictyostelium to study the functions of proteins linked to neuronal ceroid lipofuscinosis.J Biomed Sci. 2016 Nov 24;23(1):83. doi: 10.1186/s12929-016-0301-0. J Biomed Sci. 2016. PMID: 27881166 Free PMC article. Review.
-
The emerging role of the first 17 amino acids of huntingtin in Huntington's disease.Biomol Concepts. 2015 Mar;6(1):33-46. doi: 10.1515/bmc-2015-0001. Biomol Concepts. 2015. PMID: 25741791 Free PMC article. Review.
-
The evolution of the huntingtin-associated protein 40 (HAP40) in conjunction with huntingtin.BMC Evol Biol. 2020 Dec 9;20(1):162. doi: 10.1186/s12862-020-01705-5. BMC Evol Biol. 2020. PMID: 33297953 Free PMC article.
-
An evolutionary recent neuroepithelial cell adhesion function of huntingtin implicates ADAM10-Ncadherin.Nat Neurosci. 2012 May;15(5):713-21. doi: 10.1038/nn.3080. Nat Neurosci. 2012. PMID: 22466506
-
Dictyostelium discoideum: A Model System for Neurological Disorders.Cells. 2022 Jan 28;11(3):463. doi: 10.3390/cells11030463. Cells. 2022. PMID: 35159273 Free PMC article. Review.
References
-
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, et al. Neuropathological classification of Huntington's disease. J Neuropathol Exp Neurol. 1985;44(6):559–577. - PubMed
-
- Martin JB, Gusella JF. Huntington's disease: Pathogenesis and management. N Engl J Med. 1986;315(20):1267–1276. - PubMed
-
- Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306(5940):234–238. - PubMed
-
- Gusella JF, MacDonald ME. Huntington's Disease. Semin Cell Biol. 1995;6:21–28. - PubMed
-
- The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's Disease chromosomes. Cell. 1993;72:971–983. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

