In Silico Augmentation of the Drug Development Pipeline: Examples from the study of Acute Inflammation
- PMID: 21552346
- PMCID: PMC3086282
- DOI: 10.1002/ddr.20415
In Silico Augmentation of the Drug Development Pipeline: Examples from the study of Acute Inflammation
Abstract
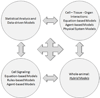
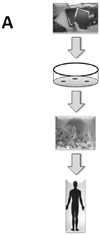
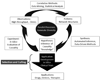
The clinical translation of promising basic biomedical findings, whether derived from reductionist studies in academic laboratories or as the product of extensive high-throughput and -content screens in the biotechnology and pharmaceutical industries, has reached a period of stagnation in which ever higher research and development costs are yielding ever fewer new drugs. Systems biology and computational modeling have been touted as potential avenues by which to break through this logjam. However, few mechanistic computational approaches are utilized in a manner that is fully cognizant of the inherent clinical realities in which the drugs developed through this ostensibly rational process will be ultimately used. In this article, we present a Translational Systems Biology approach to inflammation. This approach is based on the use of mechanistic computational modeling centered on inherent clinical applicability, namely that a unified suite of models can be applied to generate in silico clinical trials, individualized computational models as tools for personalized medicine, and rational drug and device design based on disease mechanism.
Figures

References
-
- Alt W, Lauffenburger DA. Transient behavior of a chemotaxis system modelling certain types of tissue inflammation. J.Math.Biol. 1987;24(6):691–722. - PubMed
-
- Alverdy J, Zaborina O, Wu L. The impact of stress and nutrition on bacterial-host interactions at the intestinal epithelial surface. Curr.Opin.Clin.Nutr.Metab Care. 2005;8(2):205–209. - PubMed
-
- An G. Agent-based computer simulation and SIRS: building a bridge between basic science and clinical trials. Shock. 2001;16(4):266–273. - PubMed
-
- An G. In-silico experiments of existing and hypothetical cytokine-directed clinical trials using agent based modeling. Crit Care Med. 2004;32:2050–2060. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

