Varied pathways of stage IA lung adenocarcinomas discovered by integrated gene expression analysis
- PMID: 21552421
- PMCID: PMC3088877
- DOI: 10.7150/ijbs.7.551
Varied pathways of stage IA lung adenocarcinomas discovered by integrated gene expression analysis
Abstract
Background: Discovery of the progression-associated genes and pathways in lung adenocarcinoma (LAD) has important implications in understanding the molecular mechanism of tumor development. However, few studies had been performed to focus on the changes of pathways in lung adenocarcinoma development using microarray expression profile.
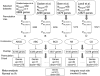
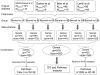
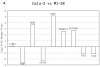
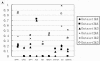
Result: We performed a meta-analysis of 4 LAD microarray datasets encompassing 353 patients to reveal differentially expressed genes (DEGs) between normal lung tissues and LAD of different stages. Overall, 1 838 genes were found to be dys-regulated, and the adipogenesis, circadian rhythm, and Id pathways were significantly changed. Interestingly, most of the genes from the same gene family (such as Interleukin receptor, Matrix metallopeptidase, Histone cluster and Minichromosome maintenance complex component families) were found to be up-regulated (or down-regulated). Real-time PCR (qRT-PCR) was applied to validate the expression of randomly selected 18 DEGs in LAD cell lines. In the pathway analysis among stages, Oxidative stress, Glycolysis/Gluconeogenesis and Integrin-mediated cell adhesion pathways, which were involved in cancer cell proliferation and metastasis, were showed to be significantly regulated in stages other than IA.
Conclusion: Genes involved in adipogenesis and Id pathways might play important roles in development of LADs. The similar trend of expression of the gene family members suggested coordinate regulation in tumor progression. Three pathways (Oxidative stress, Glycolysis/Gluconeogenesis and Integrin-mediated cell adhesion pathways) significantly regulated in stages other than stage IA suggested that genes and pathways conferring invasive character might be activated in the preinvasive stage IB, while the Oxidative stress and the Glycolysis/Gluconeogenesis pathways might have strong connections to cisplatin-based chemotherapy. The insignificantly regulated three pathways in stage IA might be used in early-stage detection of LAD.
Keywords: lung adenocarcinoma; meta-analysis; pathway; sample size.
Conflict of interest statement
Conflict of Interests: The authors have declared that no conflict of interest exists.
Figures


References
-
- Bild AH, Yao G, Chang JT, Wang Q, Potti A. et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2005;439:353–357. - PubMed
-
- Mareel M, Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol Rev. 2003;83:337–376. - PubMed
-
- Nordquist LT, Simon GR, Cantor A, Alberts WM, Bepler G. Improved survival in never-smokers vs current smokers with primary adenocarcinoma of the lung. Chest. 2004;126:347–351. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

