Inactivation of the DNA repair genes mutS, mutL or the anti-recombination gene mutS2 leads to activation of vitamin B1 biosynthesis genes
- PMID: 21552516
- PMCID: PMC3084264
- DOI: 10.1371/journal.pone.0019053
Inactivation of the DNA repair genes mutS, mutL or the anti-recombination gene mutS2 leads to activation of vitamin B1 biosynthesis genes
Abstract
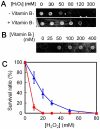
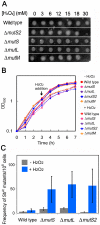
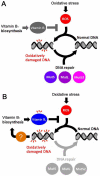
Oxidative stress generates harmful reactive oxygen species (ROS) that attack biomolecules including DNA. In living cells, there are several mechanisms for detoxifying ROS and repairing oxidatively-damaged DNA. In this study, transcriptomic analyses clarified that disruption of DNA repair genes mutS and mutL, or the anti-recombination gene mutS2, in Thermus thermophilus HB8, induces the biosynthesis pathway for vitamin B(1), which can serve as an ROS scavenger. In addition, disruption of mutS, mutL, or mutS2 resulted in an increased rate of oxidative stress-induced mutagenesis. Co-immunoprecipitation and pull-down experiments revealed previously-unknown interactions of MutS2 with MutS and MutL, indicating that these proteins cooperatively participate in the repair of oxidatively damaged DNA. These results suggested that bacterial cells sense the accumulation of oxidative DNA damage or absence of DNA repair activity, and signal the information to the transcriptional regulation machinery for an ROS-detoxifying system.
Conflict of interest statement
Figures



Similar articles
-
Thermus thermophilus MutS2, a MutS paralogue, possesses an endonuclease activity promoted by MutL.J Biochem. 2004 Mar;135(3):375-84. doi: 10.1093/jb/mvh045. J Biochem. 2004. PMID: 15113836
-
Structural and functional divergence of MutS2 from bacterial MutS1 and eukaryotic MSH4-MSH5 homologs.J Bacteriol. 2005 May;187(10):3528-37. doi: 10.1128/JB.187.10.3528-3537.2005. J Bacteriol. 2005. PMID: 15866941 Free PMC article.
-
Nuclease activity of the MutS homologue MutS2 from Thermus thermophilus is confined to the Smr domain.Nucleic Acids Res. 2007;35(3):850-60. doi: 10.1093/nar/gkl735. Epub 2007 Jan 10. Nucleic Acids Res. 2007. PMID: 17215294 Free PMC article.
-
Bacterial DNA repair genes and their eukaryotic homologues: 5. The role of recombination in DNA repair and genome stability.Acta Biochim Pol. 2007;54(3):483-94. Epub 2007 Sep 23. Acta Biochim Pol. 2007. PMID: 17893749 Review.
-
Wot the 'L-Does MutL do?Mutat Res. 2010 Dec;705(3):228-38. doi: 10.1016/j.mrrev.2010.07.002. Epub 2010 Aug 3. Mutat Res. 2010. PMID: 20667509 Review.
Cited by
-
Enhancing E. coli tolerance towards oxidative stress via engineering its global regulator cAMP receptor protein (CRP).PLoS One. 2012;7(12):e51179. doi: 10.1371/journal.pone.0051179. Epub 2012 Dec 14. PLoS One. 2012. PMID: 23251448 Free PMC article.
-
Characterization of multi-functional properties and conformational analysis of MutS2 from Thermotoga maritima MSB8.PLoS One. 2012;7(4):e34529. doi: 10.1371/journal.pone.0034529. Epub 2012 Apr 24. PLoS One. 2012. PMID: 22545085 Free PMC article.
-
Molecular basis for the functions of a bacterial MutS2 in DNA repair and recombination.DNA Repair (Amst). 2017 Sep;57:161-170. doi: 10.1016/j.dnarep.2017.07.004. Epub 2017 Jul 19. DNA Repair (Amst). 2017. PMID: 28800560 Free PMC article.
-
Expansion of the SOS regulon of Vibrio cholerae through extensive transcriptome analysis and experimental validation.BMC Genomics. 2018 May 21;19(1):373. doi: 10.1186/s12864-018-4716-8. BMC Genomics. 2018. PMID: 29783948 Free PMC article.
-
Putrescine Supplementation Limits the Expansion of pks+ Escherichia coli and Tumor Development in the Colon.Cancer Res Commun. 2024 Jul 1;4(7):1777-1792. doi: 10.1158/2767-9764.CRC-23-0355. Cancer Res Commun. 2024. PMID: 38934090 Free PMC article.
References
-
- Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat Res. 2003;531:5–23. - PubMed
-
- Jovanovic SV, Simic MG. The DNA guanyl radical: kinetics and mechanisms of generation and repair. Biochim Biophys Acta. 1989;1008:39–44. - PubMed
-
- Friedberg EC, Walker GC, Siede W. Washington DC: American Society for Microbiology; 2005. DNA repair and mutagenesis.
-
- Hsu GW, Ober M, Carell T, Beese LS. Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase. Nature. 2004;431:217–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

