Critical factors governing the difference in antizyme-binding affinities between human ornithine decarboxylase and antizyme inhibitor
- PMID: 21552531
- PMCID: PMC3084279
- DOI: 10.1371/journal.pone.0019253
Critical factors governing the difference in antizyme-binding affinities between human ornithine decarboxylase and antizyme inhibitor
Abstract
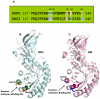
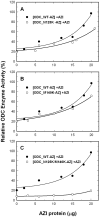
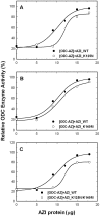
Both ornithine decarboxylase (ODC) and its regulatory protein, antizyme inhibitor (AZI), can bind with antizyme (AZ), but the latter has a higher AZ-binding affinity. The results of this study clearly identify the critical amino acid residues governing the difference in AZ-binding affinities between human ODC and AZI. Inhibition experiments using a series of ODC mutants suggested that residues 125 and 140 may be the key residues responsible for the differential AZ-binding affinities. The ODC_N125K/M140K double mutant demonstrated a significant inhibition by AZ, and the IC(50) value of this mutant was 0.08 µM, three-fold smaller than that of ODC_WT. Furthermore, the activity of the AZ-inhibited ODC_N125K/M140K enzyme was hardly rescued by AZI. The dissociation constant (K(d)) of the [ODC_N125K/M140K]-AZ heterodimer was approximately 0.02 µM, which is smaller than that of WT_ODC by approximately 10-fold and is very close to the K(d) value of AZI_WT, suggesting that ODC_N125K/M140K has an AZ-binding affinity higher than that of ODC_WT and similar to that of AZI. The efficiency of the AZI_K125N/K140M double mutant in the rescue of AZ-inhibited ODC enzyme activity was less than that of AZI_WT. The K(d) value of [AZI_K125N/K140M]-AZ was 0.18 µM, nine-fold larger than that of AZI_WT and close to the K(d) value of ODC_WT, suggesting that AZI_K125N/K140M has an AZ-binding affinity lower than that of AZI_WT and similar to that of ODC. These data support the hypothesis that the differences in residues 125 and 140 in ODC and AZI are responsible for the differential AZ-binding affinities.
Conflict of interest statement
Figures


Similar articles
-
The antizyme family for regulating polyamines.J Biol Chem. 2018 Nov 30;293(48):18730-18735. doi: 10.1074/jbc.TM118.003339. Epub 2018 Oct 24. J Biol Chem. 2018. PMID: 30355739 Free PMC article. Review.
-
Minimal antizyme peptide fully functioning in the binding and inhibition of ornithine decarboxylase and antizyme inhibitor.PLoS One. 2011;6(9):e24366. doi: 10.1371/journal.pone.0024366. Epub 2011 Sep 9. PLoS One. 2011. PMID: 21931692 Free PMC article.
-
Critical factors determining dimerization of human antizyme inhibitor.J Biol Chem. 2009 Sep 25;284(39):26768-77. doi: 10.1074/jbc.M109.007807. Epub 2009 Jul 27. J Biol Chem. 2009. PMID: 19635796 Free PMC article.
-
Degradation of antizyme inhibitor, an ornithine decarboxylase homologous protein, is ubiquitin-dependent and is inhibited by antizyme.J Biol Chem. 2004 Dec 24;279(52):54097-102. doi: 10.1074/jbc.M410234200. Epub 2004 Oct 18. J Biol Chem. 2004. PMID: 15491992
-
Degradation of ornithine decarboxylase by the 26S proteasome.Biochem Biophys Res Commun. 2000 Jan 7;267(1):1-6. doi: 10.1006/bbrc.1999.1706. Biochem Biophys Res Commun. 2000. PMID: 10623564 Review.
Cited by
-
The antizyme family for regulating polyamines.J Biol Chem. 2018 Nov 30;293(48):18730-18735. doi: 10.1074/jbc.TM118.003339. Epub 2018 Oct 24. J Biol Chem. 2018. PMID: 30355739 Free PMC article. Review.
-
Developing a novel FRET assay, targeting the binding between Antizyme-AZIN.Sci Rep. 2019 Mar 15;9(1):4632. doi: 10.1038/s41598-019-40929-4. Sci Rep. 2019. PMID: 30874587 Free PMC article.
-
Characterization of an androgen-responsive, ornithine decarboxylase-related protein in mouse kidney.Biosci Rep. 2017 Jul 12;37(4):BSR20170163. doi: 10.1042/BSR20170163. Print 2017 Aug 31. Biosci Rep. 2017. PMID: 28607032 Free PMC article.
-
Protein degradation of antizyme depends on the N-terminal degrons.Protein Sci. 2024 Nov;33(11):e5199. doi: 10.1002/pro.5199. Protein Sci. 2024. PMID: 39473024
-
Role of Antizyme Inhibitor Proteins in Cancers and Beyond.Onco Targets Ther. 2021 Jan 25;14:667-682. doi: 10.2147/OTT.S281157. eCollection 2021. Onco Targets Ther. 2021. PMID: 33531815 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

