The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components
- PMID: 21552543
- PMCID: PMC3084213
- DOI: 10.1371/journal.pgen.1002057
The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components
Abstract
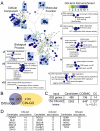
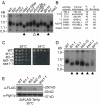
Chromosome instability (CIN) is observed in most solid tumors and is linked to somatic mutations in genome integrity maintenance genes. The spectrum of mutations that cause CIN is only partly known and it is not possible to predict a priori all pathways whose disruption might lead to CIN. To address this issue, we generated a catalogue of CIN genes and pathways by screening ∼ 2,000 reduction-of-function alleles for 90% of essential genes in Saccharomyces cerevisiae. Integrating this with published CIN phenotypes for other yeast genes generated a systematic CIN gene dataset comprised of 692 genes. Enriched gene ontology terms defined cellular CIN pathways that, together with sequence orthologs, created a list of human CIN candidate genes, which we cross-referenced to published somatic mutation databases revealing hundreds of mutated CIN candidate genes. Characterization of some poorly characterized CIN genes revealed short telomeres in mutants of the ASTRA/TTT components TTI1 and ASA1. High-throughput phenotypic profiling links ASA1 to TTT (Tel2-Tti1-Tti2) complex function and to TORC1 signaling via Tor1p stability, consistent with the role of TTT in PI3-kinase related kinase biogenesis. The comprehensive CIN gene list presented here in principle comprises all conserved eukaryotic genome integrity pathways. Deriving human CIN candidate genes from the list allows direct cross-referencing with tumor mutational data and thus candidate mutations potentially driving CIN in tumors. Overall, the CIN gene spectrum reveals new chromosome biology and will help us to understand CIN phenotypes in human disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumors. Trends Cell Biol. 1999;9:M57–60. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

