Biomarker discovery and redundancy reduction towards classification using a multi-factorial MALDI-TOF MS T2DM mouse model dataset
- PMID: 21554713
- PMCID: PMC3116487
- DOI: 10.1186/1471-2105-12-140
Biomarker discovery and redundancy reduction towards classification using a multi-factorial MALDI-TOF MS T2DM mouse model dataset
Abstract
Background: Diabetes like many diseases and biological processes is not mono-causal. On the one hand multi-factorial studies with complex experimental design are required for its comprehensive analysis. On the other hand, the data from these studies often include a substantial amount of redundancy such as proteins that are typically represented by a multitude of peptides. Coping simultaneously with both complexities (experimental and technological) makes data analysis a challenge for Bioinformatics.
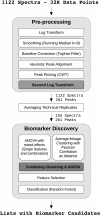
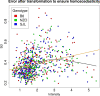
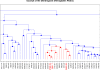
Results: We present a comprehensive work-flow tailored for analyzing complex data including data from multi-factorial studies. The developed approach aims at revealing effects caused by a distinct combination of experimental factors, in our case genotype and diet. Applying the developed work-flow to the analysis of an established polygenic mouse model for diet-induced type 2 diabetes, we found peptides with significant fold changes exclusively for the combination of a particular strain and diet. Exploitation of redundancy enables the visualization of peptide correlation and provides a natural way of feature selection for classification and prediction. Classification based on the features selected using our approach performs similar to classifications based on more complex feature selection methods.
Conclusions: The combination of ANOVA and redundancy exploitation allows for identification of biomarker candidates in multi-dimensional MALDI-TOF MS profiling studies with complex experimental design. With respect to feature selection our method provides a fast and intuitive alternative to global optimization strategies with comparable performance. The method is implemented in R and the scripts are available by contacting the corresponding author.
Figures



References
-
- Tiffin N, Adie E, Turner F, Brunner HG, van Driel MA, Oti M, Lopez-Bigas N, Ouzounis C, Perez-Iratxeta C, Andrade-Navarro MA, Adeyemo A, Patti ME, Semple CA, Hide W. Computational disease gene identification: a concert of methods prioritizes type 2 diabetes and obesity candidate genes. Nucleic Acids Res. 2006;34:3067–3081. doi: 10.1093/nar/gkl381. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

