The impact of oxygen on the transcriptome of recombinant S. cerevisiae and P. pastoris - a comparative analysis
- PMID: 21554735
- PMCID: PMC3116504
- DOI: 10.1186/1471-2164-12-218
The impact of oxygen on the transcriptome of recombinant S. cerevisiae and P. pastoris - a comparative analysis
Abstract
Background: Saccharomyces cerevisiae and Pichia pastoris are two of the most relevant microbial eukaryotic platforms for the production of recombinant proteins. Their known genome sequences enabled several transcriptomic profiling studies under many different environmental conditions, thus mimicking not only perturbations and adaptations which occur in their natural surroundings, but also in industrial processes. Notably, the majority of such transcriptome analyses were performed using non-engineered strains.In this comparative study, the gene expression profiles of S. cerevisiae and P. pastoris, a Crabtree positive and Crabtree negative yeast, respectively, were analyzed for three different oxygenation conditions (normoxic, oxygen-limited and hypoxic) under recombinant protein producing conditions in chemostat cultivations.
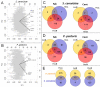
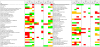
Results: The major differences in the transcriptomes of S. cerevisiae and P. pastoris were observed between hypoxic and normoxic conditions, where the availability of oxygen strongly affected ergosterol biosynthesis, central carbon metabolism and stress responses, particularly the unfolded protein response. Steady state conditions under low oxygen set-points seemed to perturb the transcriptome of S. cerevisiae to a much lesser extent than the one of P. pastoris, reflecting the major tolerance of the baker's yeast towards oxygen limitation, and a higher fermentative capacity. Further important differences were related to Fab production, which was not significantly affected by oxygen availability in S. cerevisiae, while a clear productivity increase had been previously reported for hypoxically grown P. pastoris.
Conclusions: The effect of three different levels of oxygen availability on the physiology of P. pastoris and S. cerevisiae revealed a very distinct remodelling of the transcriptional program, leading to novel insights into the different adaptive responses of Crabtree negative and positive yeasts to oxygen availability. Moreover, the application of such comparative genomic studies to recombinant hosts grown in different environments might lead to the identification of key factors for efficient protein production.
Figures


References
-
- Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodríguez-Carmona E, Baumann K, Giuliani M, Parrilli E, Branduardi P, Lang C. et al.Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

