Reducing conditions are the key for efficient production of active ribonuclease inhibitor in Escherichia coli
- PMID: 21554746
- PMCID: PMC3112386
- DOI: 10.1186/1475-2859-10-31
Reducing conditions are the key for efficient production of active ribonuclease inhibitor in Escherichia coli
Abstract
Background: The eukaryotic RNase ribonuclease/angiogenin inhibitors (RI) are a protein group distinguished by a unique structure - they are composed of hydrophobic leucine-rich repeat motifs (LRR) and contain a high amount of reduced cysteine residues. The members of this group are difficult to produce in E. coli and other recombinant hosts due to their high aggregation tendency.
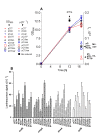
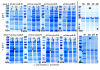
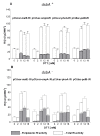
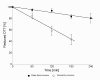
Results: In this work dithiothreitol (DTT) was successfully applied for improving the yield of correctly folded ribonuclease/angiogenin inhibitor in E. coli K12 periplasmic and cytoplasmic compartments. The feasibility of the in vivo folding concepts for cytoplasmic and periplasmic production were demonstrated at batch and fed-batch cultivation modes in shake flasks and at the bioreactor scale.Firstly, the best secretion conditions of RI in the periplasmic space were evaluated by using a high throughput multifactorial screening approach of a vector library, directly with the Enbase fed-batch production mode in 96-well plates. Secondly, the effect of the redox environment was evaluated in isogenic dsbA+ and dsbA- strains at the various cultivation conditions with reducing agents in the cultivation medium. Despite the fusion to the signal peptide, highest activities were found in the cytoplasmic fraction. Thus by removing the signal peptide the positive effect of the reducing agent DTT was clearly proven also for the cytoplasmic compartment. Finally, optimal periplasmic and cytoplasmic RI fed-batch production processes involving externally added DTT were developed in shake flasks and scaled up to the bioreactor scale.
Conclusions: DTT highly improved both, periplasmic and cytoplasmic accumulation and activity of RI at low synthesis rate, i.e. in constructs harbouring weak recombinant synthesis rate stipulating genetic elements together with cultivation at low temperature. In a stirred bioreactor environment RI folding was strongly improved by repeated pulse addition of DTT at low aeration conditions.
Figures





References
-
- Fominaya JM, Hofsteenge J. Inactivation of ribonuclease inhibitor by thiol-disulfide exchange. J Biol Chem. 1992;267:24655–24660. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

