Mining the TRAF6/p62 interactome for a selective ubiquitination motif
- PMID: 21554762
- PMCID: PMC3090762
- DOI: 10.1186/1753-6561-5-S2-S4
Mining the TRAF6/p62 interactome for a selective ubiquitination motif
Abstract
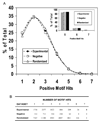
A new approach is described here to predict ubiquitinated substrates of the E3 ubiquitin ligase, TRAF6, which takes into account its interaction with the scaffold protein SQSTM1/p62. A novel TRAF6 ubiquitination motif defined as [-(hydrophobic)-k-(hydrophobic)-x-x-(hydrophobic)- (polar)-(hydrophobic)-(polar)-(hydrophobic)] was identified and used to screen the TRAF6/p62 interactome composed of 155 proteins, that were either TRAF6 or p62 interactors, or a negative dataset, composed of 54 proteins with no known association to either TRAF6 or p62. NRIF (K19), TrkA (K485), TrkB (K811), TrkC (K602 and K815), NTRK2 (K828), NTRK3 (K829) and MBP (K169) were found to possess a perfect match for the amino acid consensus motif for TRAF6/p62 ubiquitination. Subsequent analyses revealed that this motif was biased to the C-terminal regions of the protein (nearly 50% the sites), and had preference for loops (~50%) and helices (~37%) over beta-strands (15% or less). In addition, the motif was observed to be in regions that were highly solvent accessible (nearly 90%). Our findings suggest that specific Lysines may be selected for ubiquitination based upon an embedded code defined by a specific amino acid motif with structural determinants. Collectively, our results reveal an unappreciated role for the scaffold protein in targeting ubiquitination. The findings described herein could be used to aid in identification of other E3/scaffold ubiquitination sites.
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous

