miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice
- PMID: 21555486
- PMCID: PMC3173243
- DOI: 10.1084/jem.20101823
miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice
Abstract
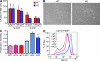
Excessive or inappropriate activation of the immune system can be deleterious to the organism, warranting multiple molecular mechanisms to control and properly terminate immune responses. MicroRNAs (miRNAs), ∼22-nt-long noncoding RNAs, have recently emerged as key posttranscriptional regulators, controlling diverse biological processes, including responses to non-self. In this study, we examine the biological role of miR-146a using genetically engineered mice and show that targeted deletion of this gene, whose expression is strongly up-regulated after immune cell maturation and/or activation, results in several immune defects. Collectively, our findings suggest that miR-146a plays a key role as a molecular brake on inflammation, myeloid cell proliferation, and oncogenic transformation.
Figures
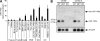





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

