Two novel CHN1 mutations in 2 families with Duane retraction syndrome
- PMID: 21555619
- PMCID: PMC3517173
- DOI: 10.1001/archophthalmol.2011.84
Two novel CHN1 mutations in 2 families with Duane retraction syndrome
Abstract
Objective: To determine the genetic cause of Duane retraction syndrome (DRS) in 2 families segregating DRS as a dominant trait.
Methods: Members of 2 unrelated pedigrees were enrolled in a genetic study. Linkage analysis was performed on the CHN1 locus. Probands and family members were screened for CHN1 mutations.
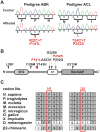
Results: The 6 affected individuals in the 2 pedigrees have DRS. Both pedigrees are consistent with linkage to the locus. Sequence analysis revealed 2 novel heterozygous missense CHN1 mutations, c.422C>T and c.754C>T, predicted to result in α2-chimaerin amino acid substitutions P141L and P252S, respectively.
Conclusions: Genetic analysis of 2 pedigrees revealed 2 novel DRS mutations, bringing the number of DRS pedigrees known to harbor CHN1 from 7 to 9. Both mutations alter residues that participate in intramolecular interactions that stabilize the inactive, closed conformation of α2-chimaerin and, thus, are predicted to result in its hyperactivation. Moreover, amino acid residue P252 was previously reported to be altered to a different residue in a previously reported DRS pedigree; thus, this is the first report of 2 CHN1 mutations altering the same residue, further supporting a gain-of-function etiology.
Clinical relevance: Members of families segregating DRS as an autosomal dominant trait should be screened for mutations in the CHN1 gene, enhancing genetic counseling and permitting earlier diagnosis.
Figures


References
-
- Engle EC. The genetic basis of complex strabismus. Pediatr Res. 2006 Mar;59(3):343–348. - PubMed
-
- DeRespinis PA, Caputo AR, Wagner RS, Guo S. Duane’s retraction syndrome. Surv Ophthalmol. 1993;38(3):258–288. - PubMed
-
- Hotchkiss MG, Miller NR, Clark AW, Green WG. Bilateral Duane’s retraction syndrome: A clinical-pathological case report. Arch Ophthalmol. 1980 May;98:870–874. - PubMed
-
- Miller NR, Kiel SM, Green WR, Clark AW. Unilateral Duane’s retraction syndrome (type 1) Arch Ophthalmol. 1982 Sep;100(9):1468–1472. - PubMed

