Exhaustion of tumor-specific CD8⁺ T cells in metastases from melanoma patients
- PMID: 21555851
- PMCID: PMC3104769
- DOI: 10.1172/JCI46102
Exhaustion of tumor-specific CD8⁺ T cells in metastases from melanoma patients
Abstract
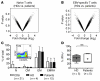
In chronic viral infections, CD8⁺ T cells become functionally deficient and display multiple molecular alterations. In contrast, only little is known of self- and tumor-specific CD8⁺ T cells from mice and humans. Here we determined molecular profiles of tumor-specific CD8⁺ T cells from melanoma patients. In peripheral blood from patients vaccinated with CpG and the melanoma antigen Melan-A/MART-1 peptide, we found functional effector T cell populations, with only small but nevertheless significant differences in T cells specific for persistent herpesviruses (EBV and CMV). In contrast, Melan-A/MART-1-specific T cells isolated from metastases from patients with melanoma expressed a large variety of genes associated with T cell exhaustion. The identified exhaustion profile revealed extended molecular alterations. Our data demonstrate a remarkable coexistence of effector cells in circulation and exhausted cells in the tumor environment. Functional T cell impairment is mediated by inhibitory receptors and further molecular pathways, which represent potential targets for cancer therapy.
Figures


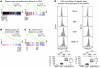


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

