Modulating F-actin organization induces organ growth by affecting the Hippo pathway
- PMID: 21556047
- PMCID: PMC3116287
- DOI: 10.1038/emboj.2011.157
Modulating F-actin organization induces organ growth by affecting the Hippo pathway
Abstract
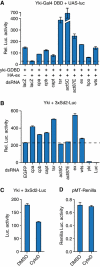
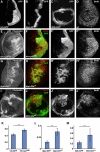
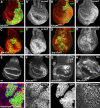
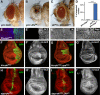
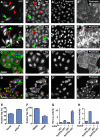
The Hippo tumour suppressor pathway is a conserved signalling pathway that controls organ size. The core of the Hpo pathway is a kinase cascade, which in Drosophila involves the Hpo and Warts kinases that negatively regulate the activity of the transcriptional coactivator Yorkie. Although several additional components of the Hippo pathway have been discovered, the inputs that regulate Hippo signalling are not fully understood. Here, we report that induction of extra F-actin formation, by loss of Capping proteins A or B, or caused by overexpression of an activated version of the formin Diaphanous, induced strong overgrowth in Drosophila imaginal discs through modulating the activity of the Hippo pathway. Importantly, loss of Capping proteins and Diaphanous overexpression did not significantly affect cell polarity and other signalling pathways, including Hedgehog and Decapentaplegic signalling. The interaction between F-actin and Hpo signalling is evolutionarily conserved, as the activity of the mammalian Yorkie-orthologue Yap is modulated by changes in F-actin. Thus, regulators of F-actin, and in particular Capping proteins, are essential for proper growth control by affecting Hippo signalling.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Bazellieres E, Assemat E, Arsanto JP, Le Bivic A, Massey-Harroche D (2009) Crumbs proteins in epithelial morphogenesis. Front Biosci 14: 2149–2169 - PubMed
-
- Benlali A, Draskovic I, Hazelett DJ, Treisman JE (2000) Act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell 101: 271–281 - PubMed
-
- Bennett FC, Harvey KF (2006) Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol 16: 2101–2110 - PubMed
-
- Boedigheimer M, Laughon A (1993) Expanded: a gene involved in the control of cell proliferation in imaginal discs. Development 118: 1291–1301 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

