Skeletal myosin light chain kinase regulates skeletal myogenesis by phosphorylation of MEF2C
- PMID: 21556048
- PMCID: PMC3116284
- DOI: 10.1038/emboj.2011.153
Skeletal myosin light chain kinase regulates skeletal myogenesis by phosphorylation of MEF2C
Abstract
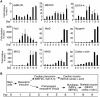
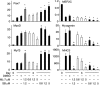
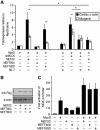
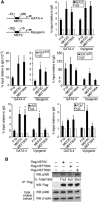
The MEF2 factors regulate transcription during cardiac and skeletal myogenesis. MEF2 factors establish skeletal muscle commitment by amplifying and synergizing with MyoD. While phosphorylation is known to regulate MEF2 function, lineage-specific regulation is unknown. Here, we show that phosphorylation of MEF2C on T(80) by skeletal myosin light chain kinase (skMLCK) enhances skeletal and not cardiac myogenesis. A phosphorylation-deficient MEF2C mutant (MEFT80A) enhanced cardiac, but not skeletal myogenesis in P19 stem cells. Further, MEFT80A was deficient in recruitment of p300 to skeletal but not cardiac muscle promoters. In gain-of-function studies, skMLCK upregulated myogenic regulatory factor (MRF) expression, leading to enhanced skeletal myogenesis in P19 cells and more efficient myogenic conversion. In loss-of-function studies, MLCK was essential for efficient MRF expression and subsequent myogenesis in embryonic stem (ES) and P19 cells as well as for proper activation of quiescent satellite cells. Thus, skMLCK regulates MRF expression by controlling the MEF2C-dependent recruitment of histone acetyltransferases to skeletal muscle promoters. This work identifies the first kinase that regulates MyoD and Myf5 expression in ES or satellite cells.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




Similar articles
-
Myocyte enhancer factor 2C and myogenin up-regulate each other's expression and induce the development of skeletal muscle in P19 cells.J Biol Chem. 2000 Jan 7;275(1):41-6. doi: 10.1074/jbc.275.1.41. J Biol Chem. 2000. PMID: 10617583
-
mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo.Mech Dev. 2003 Sep;120(9):1021-32. doi: 10.1016/s0925-4773(03)00178-3. Mech Dev. 2003. PMID: 14550531
-
Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C.Mol Cell Biol. 1997 Feb;17(2):1010-26. doi: 10.1128/MCB.17.2.1010. Mol Cell Biol. 1997. PMID: 9001254 Free PMC article.
-
Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis.Semin Cell Dev Biol. 2017 Dec;72:19-32. doi: 10.1016/j.semcdb.2017.11.011. Epub 2017 Nov 15. Semin Cell Dev Biol. 2017. PMID: 29127046 Review.
-
Skeletal muscle mass is controlled by the MRF4-MEF2 axis.Curr Opin Clin Nutr Metab Care. 2018 May;21(3):164-167. doi: 10.1097/MCO.0000000000000456. Curr Opin Clin Nutr Metab Care. 2018. PMID: 29389722 Review.
Cited by
-
Gli2 and MEF2C activate each other's expression and function synergistically during cardiomyogenesis in vitro.Nucleic Acids Res. 2012 Apr;40(8):3329-47. doi: 10.1093/nar/gkr1232. Epub 2011 Dec 22. Nucleic Acids Res. 2012. PMID: 22199256 Free PMC article.
-
Transcriptome Analysis of mRNA and lncRNA Related to Muscle Growth and Development in Gannan Yak and Jeryak.Int J Mol Sci. 2023 Nov 30;24(23):16991. doi: 10.3390/ijms242316991. Int J Mol Sci. 2023. PMID: 38069312 Free PMC article.
-
TNF-α/NF-κB mediated upregulation of Dectin-1 in hyperglycemic obesity: implications for metabolic inflammation and diabetes.J Transl Med. 2025 Apr 23;23(1):462. doi: 10.1186/s12967-025-06303-x. J Transl Med. 2025. PMID: 40270030 Free PMC article.
-
Whole genome sequencing of Gyeongbuk Araucana, a newly developed blue-egg laying chicken breed, reveals its origin and genetic characteristics.Sci Rep. 2016 May 24;6:26484. doi: 10.1038/srep26484. Sci Rep. 2016. PMID: 27215397 Free PMC article.
-
MiRNA sequencing of Embryonic Myogenesis in Chengkou Mountain Chicken.BMC Genomics. 2022 Aug 10;23(1):571. doi: 10.1186/s12864-022-08795-z. BMC Genomics. 2022. PMID: 35948880 Free PMC article.
References
-
- Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D (2008) The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics 7: 560–572 - PubMed
-
- Al-Madhoun AS, Chen YX, Haidari L, Rayner K, Gerthoffer W, McBride H, O’Brien ER (2007) The interaction and cellular localization of HSP27 and ERbeta are modulated by 17beta-estradiol and HSP27 phosphorylation. Mol Cell Endocrinol 270: 33–42 - PubMed
-
- Al-Madhoun AS, Johnsamuel J, Yan J, Ji W, Wang J, Zhuo JC, Lunato AJ, Woollard JE, Hawk AE, Cosquer GY, Blue TE, Eriksson S, Tjarks W (2002) Synthesis of a small library of 3-(carboranylalkyl)thymidines and their biological evaluation as substrates for human thymidine kinases 1 and 2. J Med Chem 45: 4018–4028 - PubMed
-
- Al-Madhoun AS, Talianidis I, Eriksson S (2004) Transcriptional regulation of the mouse deoxycytidine kinase: identification and functional analysis of nuclear protein binding sites at the proximal promoter. Biochem Pharmacol 68: 2397–2407 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

