Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts
- PMID: 21556056
- PMCID: PMC3112538
- DOI: 10.1038/ncomms1305
Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts
Abstract
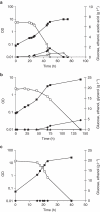
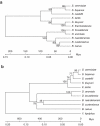
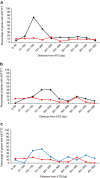
Saccharomyces yeasts degrade sugars to two-carbon components, in particular ethanol, even in the presence of excess oxygen. This characteristic is called the Crabtree effect and is the background for the 'make-accumulate-consume' life strategy, which in natural habitats helps Saccharomyces yeasts to out-compete other microorganisms. A global promoter rewiring in the Saccharomyces cerevisiae lineage, which occurred around 100 mya, was one of the main molecular events providing the background for evolution of this strategy. Here we show that the Dekkera bruxellensis lineage, which separated from the Saccharomyces yeasts more than 200 mya, also efficiently makes, accumulates and consumes ethanol and acetic acid. Analysis of promoter sequences indicates that both lineages independently underwent a massive loss of a specific cis-regulatory element from dozens of genes associated with respiration, and we show that also in D. bruxellensis this promoter rewiring contributes to the observed Crabtree effect.
Figures

References
-
- Pronk J. T., Steensma H. Y. & van Dijken J. P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12 1607–1633 (1996). - PubMed
-
- Piskur J., Rozpedowska E., Polakova S., Merico A. & Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 22 183–186 (2006). - PubMed
-
- Wolfe K. H. & Shields D. C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387 708–713 (1997). - PubMed
-
- Ihmels J. et al.. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309 938–940 (2005). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

