Stimulus-dependent dynamics of p53 in single cells
- PMID: 21556066
- PMCID: PMC3130553
- DOI: 10.1038/msb.2011.20
Stimulus-dependent dynamics of p53 in single cells
Abstract
Many biological networks respond to various inputs through a common signaling molecule that triggers distinct cellular outcomes. One potential mechanism for achieving specific input-output relationships is to trigger distinct dynamical patterns in response to different stimuli. Here we focused on the dynamics of p53, a tumor suppressor activated in response to cellular stress. We quantified the dynamics of p53 in individual cells in response to UV and observed a single pulse that increases in amplitude and duration in proportion to the UV dose. This graded response contrasts with the previously described series of fixed pulses in response to γ-radiation. We further found that while γ-triggered p53 pulses are excitable, the p53 response to UV is not excitable and depends on continuous signaling from the input-sensing kinases. Using mathematical modeling and experiments, we identified feedback loops that contribute to specific features of the stimulus-dependent dynamics of p53, including excitability and input-duration dependency. Our study shows that different stresses elicit different temporal profiles of p53, suggesting that modulation of p53 dynamics might be used to achieve specificity in this network.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
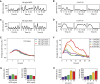
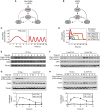

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

