Evidence for a fourteenth mtDNA-encoded protein in the female-transmitted mtDNA of marine Mussels (Bivalvia: Mytilidae)
- PMID: 21556327
- PMCID: PMC3083442
- DOI: 10.1371/journal.pone.0019365
Evidence for a fourteenth mtDNA-encoded protein in the female-transmitted mtDNA of marine Mussels (Bivalvia: Mytilidae)
Abstract
Background: A novel feature for animal mitochondrial genomes has been recently established: i.e., the presence of additional, lineage-specific, mtDNA-encoded proteins with functional significance. This feature has been observed in freshwater mussels with doubly uniparental inheritance of mtDNA (DUI). The latter unique system of mtDNA transmission, which also exists in some marine mussels and marine clams, is characterized by one mt genome inherited from the female parent (F mtDNA) and one mt genome inherited from the male parent (M mtDNA). In freshwater mussels, the novel mtDNA-encoded proteins have been shown to be mt genome-specific (i.e., one novel protein for F genomes and one novel protein for M genomes). It has been hypothesized that these novel, F- and M-specific, mtDNA-encoded proteins (and/or other F- and/or M-specific mtDNA sequences) could be responsible for the different modes of mtDNA transmission in bivalves but this remains to be demonstrated.
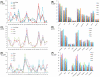
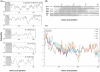
Methodology/principal findings: We investigated all complete (or nearly complete) female- and male-transmitted marine mussel mtDNAs previously sequenced for the presence of ORFs that could have functional importance in these bivalves. Our results confirm the presence of a novel F genome-specific mt ORF, of significant length (>100aa) and located in the control region, that most likely has functional significance in marine mussels. The identification of this ORF in five Mytilus species suggests that it has been maintained in the mytilid lineage (subfamily Mytilinae) for ∼13 million years. Furthermore, this ORF likely has a homologue in the F mt genome of Musculista senhousia, a DUI-containing mytilid species in the subfamily Crenellinae. We present evidence supporting the functionality of this F-specific ORF at the transcriptional, amino acid and nucleotide levels.
Conclusions/significance: Our results offer support for the hypothesis that "novel F genome-specific mitochondrial genes" are involved in key biological functions in bivalve species with DUI.
Conflict of interest statement
Figures



References
-
- Birky CW., Jr The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annu Rev Genet. 2001;35:125–148. - PubMed
-
- Elson JL, Lightowlers RN. Mitochondrial DNA clonality in the dock: can surveillance swing the case? Trends Genet. 2006;22:603–607. - PubMed
-
- Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101:301–320. - PubMed
-
- Garesse R, Vallejo CG. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene. 2001;263:1–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

