Advances in mass spectrometry for the identification of pathogens
- PMID: 21557290
- PMCID: PMC7168406
- DOI: 10.1002/mas.20320
Advances in mass spectrometry for the identification of pathogens
Abstract
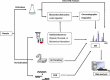
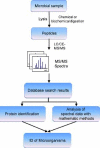
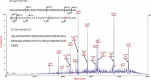
Mass spectrometry (MS) has become an important technique to identify microbial biomarkers. The rapid and accurate MS identification of microorganisms without any extensive pretreatment of samples is now possible. This review summarizes MS methods that are currently utilized in microbial analyses. Affinity methods are effective to clean, enrich, and investigate microorganisms from complex matrices. Functionalized magnetic nanoparticles might concentrate traces of target microorganisms from sample solutions. Therefore, nanoparticle-based techniques have a favorable detection limit. MS coupled with various chromatographic techniques, such as liquid chromatography and capillary electrophoresis, reduces the complexity of microbial biomarkers and yields reliable results. The direct analysis of whole pathogenic microbial cells with matrix-assisted laser desorption/ionization MS without sample separation reveals specific biomarkers for taxonomy, and has the advantages of simplicity, rapidity, and high-throughput measurements. The MS detection of polymerase chain reaction (PCR)-amplified microbial nucleic acids provides an alternative to biomarker analysis. This review will conclude with some current applications of MS in the identification of pathogens.
Copyright © 2011 Wiley Periodicals, Inc.
Figures


References
-
- Abbas‐Hawks C, Voorhees KJ, Miketova P. 2006. Identification of carbohydrate and nucleic acid biomarkers in the pyrolysis electron ionization‐high‐resolution mass spectrum of Brucella neotomae . J Anal Appl Pyrol 76: 6–13.
-
- Adams KL, Steele PT, Bogan MJ, Sadler NM, Martin SI, Martin AN, Frank M. 2008. Reagentless detection of mycobacteria tuberculosis H37RA in respiratory effluents in minutes. Anal Chem 80: 5350–5357. - PubMed
-
- Afonso C, Fenselau C. 2003. Use of bioactive glass slides for matrix‐assisted laser desorption/ionization analysis: Application to microorganisms. Anal Chem 75: 694–697. - PubMed
-
- Al Dahouk S, Nockler K, Scholz HC, Tomaso H, Bogumil R, Neubauer H. 2006. Immunoproteomic characterization of Brucella abortus 1119‐3 preparations used for the serodiagnosis of Brucella infections. J Immunol Methods 309: 34–47. - PubMed
-
- Anhalt JP, Fenselau C. 1975. Identification of bacteria using mass spectrometry. Anal Chem 47: 219–225.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

