Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7
- PMID: 21557376
- PMCID: PMC3149194
- DOI: 10.1002/pro.649
Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7
Abstract
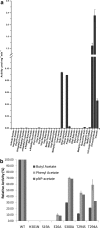
There is a high prevalence of sialic acid in a number of different organisms, resulting in there being a myriad of different enzymes that can exploit it as a fermentable carbon source. One such enzyme is NanS, a carbohydrate esterase that we show here deacetylates the 9 position of 9-O-sialic acid so that it can be readily transported into the cell for catabolism. Through structural studies, we show that NanS adopts a SGNH hydrolase fold. Although the backbone of the structure is similar to previously characterized family members, sequence comparisons indicate that this family can be further subdivided into two subfamilies with somewhat different fingerprints. NanS is the founding member of group II. Its catalytic center contains Ser19 and His301 but no Asp/Glu is present to form the classical catalytic triad. The contribution of Ser19 and His301 to catalysis was confirmed by mutagenesis. In addition to structural characterization, we have mapped the specificity of NanS using a battery of substrates.
Copyright © 2011 The Protein Society.
Figures
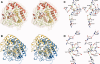

References
-
- Williamson G. Purification and characterization of pectin acetylesterase from orange peel. Phytochemistry. 1991;30:445–449.
-
- Sundberg M, Poutanen K. Purification and properties of two acetylxylan esterases of Trichoderma reesei. Biotech Appl Biochem. 1991;13:1–11.
-
- Tenkanen M, Thornton J, Viikari L. An acetylglucomannan esterase of Aspergillus oryzae; purification, characterization and role in the hydrolysis of O-acetyl-galactoglucomannan. J Biotechnol. 1995;42:197–206. - PubMed
-
- Araki Y, Ito E. A pathway of chitosan formation in Mucor rouxii: enzymatic deacetylation of chitin. Biochem Biophys Res Commun. 1974;56:669–675. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

