Identification and characterization of the Chlamydia trachomatis L2 S-adenosylmethionine transporter
- PMID: 21558433
- PMCID: PMC3104491
- DOI: 10.1128/mBio.00051-11
Identification and characterization of the Chlamydia trachomatis L2 S-adenosylmethionine transporter
Abstract
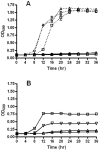
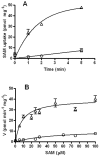
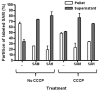
Methylation is essential to the physiology of all cells, including the obligate intracellular bacterium Chlamydia. Nevertheless, the methylation cycle is under strong reductive evolutionary pressure in Chlamydia. Only Parachlamydia acanthamoebae and Waddlia chondrophila genome sequences harbor homologs to metK, encoding the S-adenosylmethionine (SAM) synthetase required for synthesis of SAM, and to sahH, which encodes the S-adenosylhomocysteine (SAH) hydrolase required for detoxification of SAH formed after the transfer of the methyl group from SAM to the methylation substrate. Transformation of a conditional-lethal ΔmetK mutant of Escherichia coli with a genomic library of Chlamydia trachomatis L2 identified CTL843 as a putative SAM transporter based on its ability to allow the mutant to survive metK deficiency only in the presence of extracellular SAM. CTL843 belongs to the drug/metabolite superfamily of transporters and allowed E. coli to transport S-adenosyl-L-[methyl-(14)C]methionine with an apparent K(m) of 5.9 µM and a V(max) of 32 pmol min(-1) mg(-1). Moreover, CTL843 conferred a growth advantage to a Δpfs E. coli mutant that lost the ability to detoxify SAH, while competition and back-transport experiments further implied that SAH was an additional substrate for CTL843. We propose that CTL843 acts as a SAM/SAH transporter (SAMHT) serving a dual function by allowing Chlamydia to acquire SAM from the host cell and excrete the toxic by-product SAH. The demonstration of a functional SAMHT provides further insight into the reductive evolution associated with the obligate intracellular lifestyle of Chlamydia and identifies an excellent chemotherapeutic target.
Importance: Obligate intracellular parasites like Chlamydia have followed a reductive evolutionary path that has made them almost totally dependent on their host cell for nutrients. In this work, we identify a unique transporter of a metabolite essential for all methylation reactions that potentially bypasses the need for two enzymatic reactions in Chlamydia. The transporter, CTL843, allows Chlamydia trachomatis L2 to steal S-adenosylmethionine (SAM) from the eukaryotic host cytosol and to likely remove the toxic S-adenosylhomocysteine (SAH) formed when SAM loses its methyl group, acting as a SAM/SAH transporter (SAMHT). In addition to reflecting the adaptation of Chlamydia to an obligate intracellular lifestyle, the specific and central roles of SAMHT in Chlamydia metabolism provide a target for the development of therapeutic agents for the treatment of chlamydial infections.
Figures
References
-
- Stephens RS, et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759 - PubMed
-
- Andersson JO, Andersson SG. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664–671 - PubMed
-
- Haferkamp I, et al. 2004. A candidate NAD+ transporter in an intracellular bacterial symbiont related to Chlamydiae. Nature 432:622–625 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
