Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum
- PMID: 21558492
- PMCID: PMC3149090
- DOI: 10.1074/mcp.M110.006411
Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum
Abstract
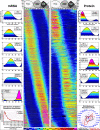
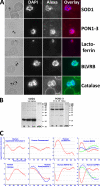
Studies of the Plasmodium falciparum transcriptome have shown that the tightly controlled progression of the parasite through the intra-erythrocytic developmental cycle (IDC) is accompanied by a continuous gene expression cascade in which most expressed genes exhibit a single transcriptional peak. Because the biochemical and cellular functions of most genes are mediated by the encoded proteins, understanding the relationship between mRNA and protein levels is crucial for inferring biological activity from transcriptional gene expression data. Although studies on other organisms show that <50% of protein abundance variation may be attributable to corresponding mRNA levels, the situation in Plasmodium is further complicated by the dynamic nature of the cyclic gene expression cascade. In this study, we simultaneously determined mRNA and protein abundance profiles for P. falciparum parasites during the IDC at 2-hour resolution based on oligonucleotide microarrays and two-dimensional differential gel electrophoresis protein gels. We find that most proteins are represented by more than one isoform, presumably because of post-translational modifications. Like transcripts, most proteins exhibit cyclic abundance profiles with one peak during the IDC, whereas the presence of functionally related proteins is highly correlated. In contrast, the abundance of most parasite proteins peaks significantly later (median 11 h) than the corresponding transcripts and often decreases slowly in the second half of the IDC. Computational modeling indicates that the considerable and varied incongruence between transcript and protein abundance may largely be caused by the dynamics of translation and protein degradation. Furthermore, we present cyclic abundance profiles also for parasite-associated human proteins and confirm the presence of five human proteins with a potential role in antioxidant defense within the parasites. Together, our data provide fundamental insights into transcript-protein relationships in P. falciparum that are important for the correct interpretation of transcriptional data and that may facilitate the improvement and development of malaria diagnostics and drug therapy.
Figures



References
-
- WHO (2009) World Health Organization, Geneva, World Malaria Report 2009
-
- Bozdech Z., Mok S., Hu G., Imwong M., Jaidee A., Russell B., Ginsburg H., Nosten F., Day N. P., White N. J., Carlton J. M., Preiser P. R. (2008) The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 105, 16290–16295 - PMC - PubMed
-
- Le Roch K. G., Zhou Y., Blair P. L., Grainger M., Moch J. K., Haynes J. D., De La Vega P., Holder A. A., Batalov S., Carucci D. J., Winzeler E. A. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301, 1503–1508 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

