Isolation and culture of larval cells from C. elegans
- PMID: 21559335
- PMCID: PMC3084877
- DOI: 10.1371/journal.pone.0019505
Isolation and culture of larval cells from C. elegans
Abstract
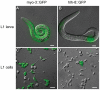
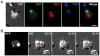
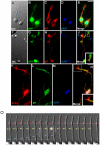
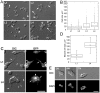
Cell culture is an essential tool to study cell function. In C. elegans the ability to isolate and culture cells has been limited to embryonically derived cells. However, cells or blastomeres isolated from mixed stage embryos terminally differentiate within 24 hours of culture, thus precluding post-embryonic stage cell culture. We have developed an efficient and technically simple method for large-scale isolation and primary culture of larval-stage cells. We have optimized the treatment to maximize cell number and minimize cell death for each of the four larval stages. We obtained up to 7.8×10(4) cells per microliter of packed larvae, and up to 97% of adherent cells isolated by this method were viable for at least 16 hours. Cultured larval cells showed stage-specific increases in both cell size and multinuclearity and expressed lineage- and cell type-specific reporters. The majority (81%) of larval cells isolated by our method were muscle cells that exhibited stage-specific phenotypes. L1 muscle cells developed 1 to 2 wide cytoplasmic processes, while L4 muscle cells developed 4 to 14 processes of various thicknesses. L4 muscle cells developed bands of myosin heavy chain A thick filaments at the cell center and spontaneously contracted ex vivo. Neurons constituted less than 10% of the isolated cells and the majority of neurons developed one or more long, microtubule-rich protrusions that terminated in actin-rich growth cones. In addition to cells such as muscle and neuron that are high abundance in vivo, we were also able to isolate M-lineage cells that constitute less than 0.2% of cells in vivo. Our novel method of cell isolation extends C. elegans cell culture to larval developmental stages, and allows use of the wealth of cell culture tools, such as cell sorting, electrophysiology, co-culture, and high-resolution imaging of subcellular dynamics, in investigation of post-embryonic development and physiology.
Conflict of interest statement
Figures

References
-
- Cowan AE, McIntosh JR. Mapping the distribution of differentiation potential for intestine, muscle, and hypodermis during early development in Caenorhabditis elegans. Cell. 1985;41:923–932. - PubMed
-
- Edgar LG, McGhee JD. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell. 1988;53:589–599. - PubMed
-
- Goldstein B. Induction of gut in Caenorhabditis elegans embryos. Nature. 1992;357:255–257. - PubMed
-
- Bloom L. Genetic and molecular analysis of genes required for axon outgrowth in caenorhabditis elegans. 1993. PhD dissertation to Massachusetts Institute of Technology.
-
- Christensen M, Estevez A, Yin X, Fox R, Morrison R, et al. A primary culture system for functional analysis of C. elegans neurons and muscle cells. Neuron. 2002;33:503–514. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

