Deep mutational scanning: assessing protein function on a massive scale
- PMID: 21561674
- PMCID: PMC3159719
- DOI: 10.1016/j.tibtech.2011.04.003
Deep mutational scanning: assessing protein function on a massive scale
Abstract
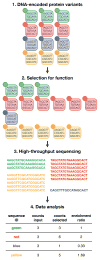
Analysis of protein mutants is an effective means to understand their function. Protein display is an approach that allows large numbers of mutants of a protein to be selected based on their activity, but only a handful with maximal activity have been traditionally identified for subsequent functional analysis. However, the recent application of high-throughput sequencing (HTS) to protein display and selection has enabled simultaneous assessment of the function of hundreds of thousands of mutants that span the activity range from high to low. Such deep mutational scanning approaches are rapid and inexpensive with the potential for broad utility. In this review, we discuss the emergence of deep mutational scanning, the challenges associated with its use and some of its exciting applications.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Olivier M, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. - PubMed
-
- Pal G, et al. Comprehensive and quantitative mapping of energy landscapes for protein-protein interactions by rapid combinatorial scanning. J Biol Chem. 2006;281:22378–22385. - PubMed
-
- Matouschek A, et al. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340:122–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

