Kinetic mechanism of human DNA ligase I reveals magnesium-dependent changes in the rate-limiting step that compromise ligation efficiency
- PMID: 21561855
- PMCID: PMC3123073
- DOI: 10.1074/jbc.M111.248831
Kinetic mechanism of human DNA ligase I reveals magnesium-dependent changes in the rate-limiting step that compromise ligation efficiency
Abstract
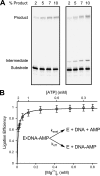
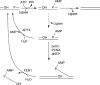
DNA ligase I (LIG1) catalyzes the ligation of single-strand breaks to complete DNA replication and repair. The energy of ATP is used to form a new phosphodiester bond in DNA via a reaction mechanism that involves three distinct chemical steps: enzyme adenylylation, adenylyl transfer to DNA, and nick sealing. We used steady state and pre-steady state kinetics to characterize the minimal mechanism for DNA ligation catalyzed by human LIG1. The ATP dependence of the reaction indicates that LIG1 requires multiple Mg(2+) ions for catalysis and that an essential Mg(2+) ion binds more tightly to ATP than to the enzyme. Further dissection of the magnesium ion dependence of individual reaction steps revealed that the affinity for Mg(2+) changes along the reaction coordinate. At saturating concentrations of ATP and Mg(2+) ions, the three chemical steps occur at similar rates, and the efficiency of ligation is high. However, under conditions of limiting Mg(2+), the nick-sealing step becomes rate-limiting, and the adenylylated DNA intermediate is prematurely released into solution. Subsequent adenylylation of enzyme prevents rebinding to the adenylylated DNA intermediate comprising an Achilles' heel of LIG1. These ligase-generated 5'-adenylylated nicks constitute persistent breaks that are a threat to genomic stability if they are not repaired. The kinetic and thermodynamic framework that we have determined for LIG1 provides a starting point for understanding the mechanism and specificity of mammalian DNA ligases.
Figures
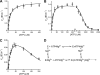
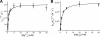

Similar articles
-
Kinetic analyses of single-stranded break repair by human DNA ligase III isoforms reveal biochemical differences from DNA ligase I.J Biol Chem. 2017 Sep 22;292(38):15870-15879. doi: 10.1074/jbc.M117.804625. Epub 2017 Jul 27. J Biol Chem. 2017. PMID: 28751376 Free PMC article.
-
Kinetics and thermodynamics of nick sealing by T4 DNA ligase.Eur J Biochem. 2003 Nov;270(21):4315-25. doi: 10.1046/j.1432-1033.2003.03824.x. Eur J Biochem. 2003. PMID: 14622296
-
Kinetic analysis of DNA strand joining by Chlorella virus DNA ligase and the role of nucleotidyltransferase motif VI in ligase adenylylation.J Biol Chem. 2012 Aug 17;287(34):28609-18. doi: 10.1074/jbc.M112.380428. Epub 2012 Jun 28. J Biol Chem. 2012. PMID: 22745124 Free PMC article.
-
Alternative Okazaki Fragment Ligation Pathway by DNA Ligase III.Genes (Basel). 2015 Jun 23;6(2):385-98. doi: 10.3390/genes6020385. Genes (Basel). 2015. PMID: 26110316 Free PMC article. Review.
-
DNA ligase I, the replicative DNA ligase.Subcell Biochem. 2012;62:327-41. doi: 10.1007/978-94-007-4572-8_17. Subcell Biochem. 2012. PMID: 22918593 Free PMC article. Review.
Cited by
-
Kinetic analyses of single-stranded break repair by human DNA ligase III isoforms reveal biochemical differences from DNA ligase I.J Biol Chem. 2017 Sep 22;292(38):15870-15879. doi: 10.1074/jbc.M117.804625. Epub 2017 Jul 27. J Biol Chem. 2017. PMID: 28751376 Free PMC article.
-
Obstacles and opportunities for base excision repair in chromatin.DNA Repair (Amst). 2022 Aug;116:103345. doi: 10.1016/j.dnarep.2022.103345. Epub 2022 May 28. DNA Repair (Amst). 2022. PMID: 35689883 Free PMC article. Review.
-
The Inhibitory Effect of Non-Substrate and Substrate DNA on the Ligation and Self-Adenylylation Reactions Catalyzed by T4 DNA Ligase.PLoS One. 2016 Mar 8;11(3):e0150802. doi: 10.1371/journal.pone.0150802. eCollection 2016. PLoS One. 2016. PMID: 26954034 Free PMC article.
-
Probing the mechanism of nick searching by LIG1 at the single-molecule level.Nucleic Acids Res. 2024 Nov 11;52(20):12604-12615. doi: 10.1093/nar/gkae865. Nucleic Acids Res. 2024. PMID: 39404052 Free PMC article.
-
Two-tiered enforcement of high-fidelity DNA ligation.Nat Commun. 2019 Nov 28;10(1):5431. doi: 10.1038/s41467-019-13478-7. Nat Commun. 2019. PMID: 31780661 Free PMC article.
References
-
- Barnes D. E., Stamp G., Rosewell I., Denzel A., Lindahl T. (1998) Curr. Biol. 8, 1395–1398 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

