Anaphase promoting complex-dependent degradation of transcriptional repressors Nrm1 and Yhp1 in Saccharomyces cerevisiae
- PMID: 21562221
- PMCID: PMC3128521
- DOI: 10.1091/mbc.E11-01-0031
Anaphase promoting complex-dependent degradation of transcriptional repressors Nrm1 and Yhp1 in Saccharomyces cerevisiae
Abstract
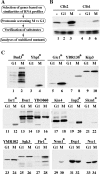
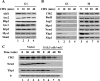
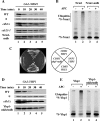
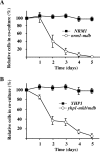
The anaphase-promoting complex/cyclosome (APC/C) is an essential ubiquitin ligase that targets cell cycle proteins for proteasome-mediated degradation in mitosis and G1. The APC regulates a number of cell cycle processes, including spindle assembly, mitotic exit, and cytokinesis, but the full range of its functions is still unknown. To better understand cellular pathways controlled by the APC, we performed a proteomic screen to identify additional APC substrates. We analyzed cell cycle-regulated proteins whose expression peaked during the period when other APC substrates were expressed. Subsequent analysis identified several proteins, including the transcriptional repressors Nrm1 and Yhp1, as authentic APC substrates. We found that APC(Cdh1) targeted Nrm1 and Yhp1 for degradation in early G1 through Destruction-box motifs and that the degradation of these repressors coincided with transcriptional activation of MBF and Mcm1 target genes, respectively. In addition, Nrm1 was stabilized by phosphorylation, most likely by the budding yeast cyclin-dependent protein kinase, Cdc28. We found that expression of stabilized forms of Nrm1 and Yhp1 resulted in reduced cell fitness, due at least in part to incomplete activation of G1-specific genes. Therefore, in addition to its known functions, APC-mediated targeting of Nrm1 and Yhp1 coordinates transcription of multiple genes in G1 with other cell cycle events.
Figures



Similar articles
-
Cdc20 protein contains a destruction-box but, unlike Clb2, its proteolysisis not acutely dependent on the activity of anaphase-promoting complex.Eur J Biochem. 2000 Jan;267(2):434-49. doi: 10.1046/j.1432-1327.2000.01014.x. Eur J Biochem. 2000. PMID: 10632713
-
Modulation of the mitotic regulatory network by APC-dependent destruction of the Cdh1 inhibitor Acm1.Mol Cell. 2008 May 23;30(4):437-46. doi: 10.1016/j.molcel.2008.04.004. Mol Cell. 2008. PMID: 18498748 Free PMC article.
-
Timely activation of budding yeast APCCdh1 involves degradation of its inhibitor, Acm1, by an unconventional proteolytic mechanism.PLoS One. 2014 Jul 29;9(7):e103517. doi: 10.1371/journal.pone.0103517. eCollection 2014. PLoS One. 2014. PMID: 25072887 Free PMC article.
-
Don't skip the G1 phase: how APC/CCdh1 keeps SCFSKP2 in check.Cell Cycle. 2004 Jul;3(7):850-2. Epub 2004 Jul 14. Cell Cycle. 2004. PMID: 15190201 Review.
-
Subunits and substrates of the anaphase-promoting complex.Exp Cell Res. 1999 May 1;248(2):339-49. doi: 10.1006/excr.1999.4443. Exp Cell Res. 1999. PMID: 10222126 Review.
Cited by
-
Identification of She3 as an SCF(Grr1) substrate in budding yeast.PLoS One. 2012;7(10):e48020. doi: 10.1371/journal.pone.0048020. Epub 2012 Oct 29. PLoS One. 2012. PMID: 23144720 Free PMC article.
-
A comprehensive, mechanistically detailed, and executable model of the cell division cycle in Saccharomyces cerevisiae.Nat Commun. 2019 Mar 21;10(1):1308. doi: 10.1038/s41467-019-08903-w. Nat Commun. 2019. PMID: 30899000 Free PMC article.
-
Cell cycle regulated transcription: from yeast to cancer.F1000Res. 2016 May 12;5:F1000 Faculty Rev-877. doi: 10.12688/f1000research.8111.1. eCollection 2016. F1000Res. 2016. PMID: 27239285 Free PMC article. Review.
-
Identification of anaphase promoting complex substrates in S. cerevisiae.PLoS One. 2012;7(9):e45895. doi: 10.1371/journal.pone.0045895. Epub 2012 Sep 26. PLoS One. 2012. PMID: 23049888 Free PMC article.
-
Create, activate, destroy, repeat: Cdk1 controls proliferation by limiting transcription factor activity.Curr Genet. 2016 May;62(2):271-6. doi: 10.1007/s00294-015-0535-5. Epub 2015 Nov 21. Curr Genet. 2016. PMID: 26590602 Review.
References
-
- Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

