Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11
- PMID: 21565612
- PMCID: PMC3261725
- DOI: 10.1016/j.cell.2011.03.041
Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11
Erratum in
- Cell. 2011 Jun 10;145(6):993
Abstract
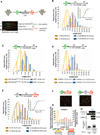
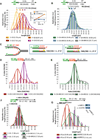
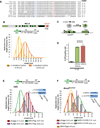
Breast cancer suppressor BRCA2 is critical for maintenance of genomic integrity and resistance to agents that damage DNA or collapse replication forks, presumably through homology-directed repair of double-strand breaks (HDR). Using single-molecule DNA fiber analysis, we show here that nascent replication tracts created before fork stalling with hydroxyurea are degraded in the absence of BRCA2 but are stable in wild-type cells. BRCA2 mutational analysis reveals that a conserved C-terminal site involved in stabilizing RAD51 filaments, but not in loading RAD51 onto DNA, is essential for this fork protection but dispensable for HDR. RAD51 filament disruption in wild-type cells phenocopies BRCA2 deficiency. BRCA2 prevents chromosomal aberrations on replication stalling, which are alleviated by inhibition of MRE11, the nuclease responsible for this form of fork instability. Thus, BRCA2 prevents rather than repairs nucleolytic lesions at stalled replication forks to maintain genomic integrity and hence likely suppresses tumorigenesis through this replication-specific function.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
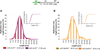


Comment in
-
DNA repair: BRCA2 gets protective at forks.Nat Rev Mol Cell Biol. 2011 Jun 2;12(7):400. doi: 10.1038/nrm3135. Nat Rev Mol Cell Biol. 2011. PMID: 21633386 No abstract available.
References
-
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. - PubMed
-
- Budzowska M, Kanaar R. Mechanisms of Dealing with DNA Damage-Induced Replication Problems. Cell Biochemistry and Biophysics. 2009;53:17–31. - PubMed
-
- Cheng WH, von Kobbe C, Opresko PL, Arthur LM, Komatsu K, Seidman MM, Carney JP, Bohr VA. Linkage between Werner syndrome protein and the Mre11 complex via Nbs1. J Biol Chem. 2004;279:21169–21176. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

