Molecular basis of secretin docking to its intact receptor using multiple photolabile probes distributed throughout the pharmacophore
- PMID: 21566140
- PMCID: PMC3129170
- DOI: 10.1074/jbc.M111.245969
Molecular basis of secretin docking to its intact receptor using multiple photolabile probes distributed throughout the pharmacophore
Abstract
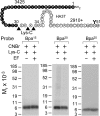
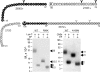
The molecular basis of ligand binding and activation of family B G protein-coupled receptors is not yet clear due to the lack of insight into the structure of intact receptors. Although NMR and crystal structures of amino-terminal domains of several family members support consistency in general structural motifs that include a peptide-binding cleft, there are variations in the details of docking of the carboxyl terminus of peptide ligands within this cleft, and there is no information about siting of the amino terminus of these peptides. There are also no empirical data to orient the receptor amino terminus relative to the core helical bundle domain. Here, we prepared a series of five new probes, incorporating photolabile moieties into positions 2, 15, 20, 24, and 25 of full agonist secretin analogues. Each bound specifically to the receptor and covalently labeled single distinct receptor residues. Peptide mapping of labeled wild-type and mutant receptors identified that the position 15, 20, and 25 probes labeled residues within the distal amino terminus of the receptor, whereas the position 24 probe labeled the amino terminus adjacent to TM1. Of note, the position 2 probe labeled a residue within the first extracellular loop of the receptor, a region not previously labeled, providing an important new constraint for docking the amino-terminal region of secretin to its receptor core. These additional experimentally derived constraints help to refine our understanding of the structure of the secretin-intact receptor complex and provide new insights into understanding the molecular mechanism for activation of family B G protein-coupled receptors.
Figures
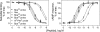





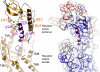

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

