The genomic architecture of sporadic heart failure
- PMID: 21566223
- PMCID: PMC3110763
- DOI: 10.1161/CIRCRESAHA.110.229260
The genomic architecture of sporadic heart failure
Abstract
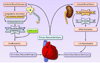
Common or sporadic systolic heart failure (heart failure) is the clinical syndrome of insufficient forward cardiac output resulting from myocardial disease. Most heart failure is the consequence of ischemic or idiopathic cardiomyopathy. There is a clear familial predisposition to heart failure, with a genetic component estimated to confer between 20% and 30% of overall risk. The multifactorial etiology of this syndrome has complicated identification of its genetic underpinnings. Until recently, almost all genetic studies of heart failure were designed and deployed according to the common disease-common variant hypothesis, in which individual risk alleles impart a small positive or negative effect and overall genetic risk is the cumulative impact of all functional genetic variations. Early studies used a candidate gene approach focused mainly on factors within adrenergic and renin-angiotensin pathways that affect heart failure progression and are targeted by standard pharmacotherapeutics. Many of these reported allelic associations with heart failure have not been replicated. However, the preponderance of data supports risk-modifier effects for the Arg389Gly polymorphism of β1-adrenergic receptors and the intron 16 in/del polymorphism of angiotensin-converting enzyme. Recent unbiased studies using genome-wide single nucleotide polymorphism microarrays have shown fewer positive results than when these platforms were applied to hypertension, myocardial infarction, or diabetes, possibly reflecting the complex etiology of heart failure. A new cardiovascular gene-centric subgenome single nucleotide polymorphism array identified a common heat failure risk allele at 1p36 in multiple independent cohorts, but the biological mechanism for this association is still uncertain. It is likely that common gene polymorphisms account for only a fraction of individual genetic heart failure risk, and future studies using deep resequencing are likely to identify rare gene variants with larger biological effects.
Figures


Similar articles
-
Genetics of common forms of heart failure.Curr Opin Cardiol. 2011 May;26(3):204-8. doi: 10.1097/HCO.0b013e328345d336. Curr Opin Cardiol. 2011. PMID: 21464711 Review.
-
Pharmacogenetics of heart failure: evidence, opportunities, and challenges for cardiovascular pharmacogenomics.J Cardiovasc Transl Res. 2008 Mar;1(1):25-36. doi: 10.1007/s12265-007-9007-8. Epub 2008 Jan 29. J Cardiovasc Transl Res. 2008. PMID: 20559955 Review.
-
Association between β1 adrenergic receptor gene Arg389Gly polymorphism and risk of heart failure: a meta-analysis.Genet Mol Res. 2015 Jun 1;14(2):5922-9. doi: 10.4238/2015.June.1.9. Genet Mol Res. 2015. PMID: 26125791
-
Common variants in HSPB7 and FRMD4B associated with advanced heart failure.Circ Cardiovasc Genet. 2010 Apr;3(2):147-54. doi: 10.1161/CIRCGENETICS.109.898395. Epub 2010 Feb 2. Circ Cardiovasc Genet. 2010. PMID: 20124441 Free PMC article.
-
Association of beta-adrenergic receptor polymorphisms and progression to heart failure in patients with idiopathic dilated cardiomyopathy.Am J Med. 2004 Oct 1;117(7):451-8. doi: 10.1016/j.amjmed.2004.04.012. Am J Med. 2004. PMID: 15464701
Cited by
-
Application of Single-Nucleotide Polymorphism-Related Risk Estimates in Identification of Increased Genetic Susceptibility to Cardiovascular Diseases: A Literature Review.Front Public Health. 2018 Jan 31;5:358. doi: 10.3389/fpubh.2017.00358. eCollection 2017. Front Public Health. 2018. PMID: 29445720 Free PMC article. Review.
-
Cardiac dyssynchrony and response to cardiac resynchronisation therapy in heart failure: can genetic predisposition play a role?Neth Heart J. 2016 Jan;24(1):11-5. doi: 10.1007/s12471-015-0766-6. Neth Heart J. 2016. PMID: 26645708 Free PMC article.
-
Familial Mortality Risks in Patients With Heart Failure-A Swedish Sibling Study.J Am Heart Assoc. 2018 Dec 18;7(24):e010181. doi: 10.1161/JAHA.118.010181. J Am Heart Assoc. 2018. PMID: 30561269 Free PMC article.
-
MicroRNAs in heart failure: Small molecules with major impact.Glob Cardiol Sci Pract. 2014 Jun 18;2014(2):79-102. doi: 10.5339/gcsp.2014.30. eCollection 2014. Glob Cardiol Sci Pract. 2014. PMID: 25419522 Free PMC article. Review.
-
Exploring the Role of Genetic and Genomic Factors in Therapeutic Response to Heart Failure: A Comprehensive Analytical Review.Genes (Basel). 2025 Jul 4;16(7):801. doi: 10.3390/genes16070801. Genes (Basel). 2025. PMID: 40725457 Free PMC article. Review.
References
-
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, Lin Y, MacDonald JR, Pang AW, Shago M, Stockwell TB, Tsiamouri A, Bafna V, Bansal V, Kravitz SA, Busam DA, Beeson KY, McIntosh TC, Remington KA, Abril JF, Gill J, Borman J, Rogers YH, Frazier ME, Scherer SW, Strausberg RL, Venter JC. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. - PMC - PubMed
-
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. - PubMed
-
- Ho CY, Seidman CE. A contemporary approach to hypertrophic cardiomyopathy. Circulation. 2006;113:e858–e862. - PubMed
-
- Judge DP. Use of genetics in the clinical evaluation of cardiomyopathy. JAMA. 2009;302:2471–2476. - PubMed
-
- Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, Ackerman MJ. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–1910. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

