Codon usage: nature's roadmap to expression and folding of proteins
- PMID: 21567958
- PMCID: PMC3166658
- DOI: 10.1002/biot.201000332
Codon usage: nature's roadmap to expression and folding of proteins
Abstract
Biomedical and biotechnological research relies on processes leading to the successful expression and production of key biological products. High-quality proteins are required for many purposes, including protein structural and functional studies. Protein expression is the culmination of multistep processes involving regulation at the level of transcription, mRNA turnover, protein translation, and post-translational modifications leading to the formation of a stable product. Although significant strides have been achieved over the past decade, advances toward integrating genomic and proteomic information are essential, and until such time, many target genes and their products may not be fully realized. Thus, the focus of this review is to provide some experimental support and a brief overview of how codon usage bias has evolved relative to regulating gene expression levels.
Copyright © 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

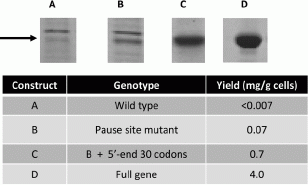
References
-
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. - PubMed
-
- Marin M. Folding at the rhythm of the rare codon beat. Biotechnol. J. 2008;3:1047–1057. - PubMed
-
- Zull JE, Smith SK. Is genetic code redundancy related to retention of structural information in both DNA strands? Trends Biochem. Sci. 1990;15:257–261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

