Growth rate regulation in Escherichia coli
- PMID: 21569058
- PMCID: PMC3478676
- DOI: 10.1111/j.1574-6976.2011.00279.x
Growth rate regulation in Escherichia coli
Abstract
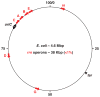
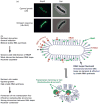
Growth rate regulation in bacteria has been an important issue in bacterial physiology for the past 50 years. This review, using Escherichia coli as a paradigm, summarizes the mechanisms for the regulation of rRNA synthesis in the context of systems biology, particularly, in the context of genome-wide competition for limited RNA polymerase (RNAP) in the cell under different growth conditions including nutrient starvation. The specific location of the seven rrn operons in the chromosome and the unique properties of the rrn promoters contribute to growth rate regulation. The length of the rrn transcripts, coupled with gene dosage effects, influence the distribution of RNAP on the chromosome in response to growth rate. Regulation of rRNA synthesis depends on multiple factors that affect the structure of the nucleoid and the allocation of RNAP for global gene expression. The magic spot ppGpp, which acts with DksA synergistically, is a key effector in both the growth rate regulation and the stringent response induced by nutrient starvation, mainly because the ppGpp level changes in response to environmental cues. It regulates rRNA synthesis via a cascade of events including both transcription initiation and elongation, and can be explained by an RNAP redistribution (allocation) model.
FEMS Microbiology Reviews © 2011 Federation of European Microbiological Societies. Published by Blackwell Publishing Ltd. No claim to original US government works.
Figures


References
-
- Adhya S, Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982;29:939–944. - PubMed
-
- Afflerbach H, Schroder O, Wagner R. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol Microbiol. 1998;28:641–653. - PubMed
-
- Aiyar SE, McLeod SM, Ross W, Hirvonen CA, Thomas MS, Johnson RC, Gourse RL. Architecture of Fis-activated transcription complexes at the Escherichia coli rrnB P1 and rrnE P1 promoters. J Mol Biol. 2002;316:501–516. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

