The midgut transcriptome of Phlebotomus (Larroussius) perniciosus, a vector of Leishmania infantum: comparison of sugar fed and blood fed sand flies
- PMID: 21569254
- PMCID: PMC3107814
- DOI: 10.1186/1471-2164-12-223
The midgut transcriptome of Phlebotomus (Larroussius) perniciosus, a vector of Leishmania infantum: comparison of sugar fed and blood fed sand flies
Abstract
Background: Parasite-vector interactions are fundamental in the transmission of vector-borne diseases such as leishmaniasis. Leishmania development in the vector sand fly is confined to the digestive tract, where sand fly midgut molecules interact with the parasites. In this work we sequenced and analyzed two midgut-specific cDNA libraries from sugar fed and blood fed female Phlebotomus perniciosus and compared the transcript expression profiles.
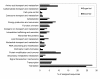
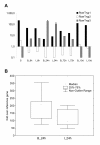
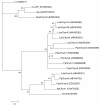
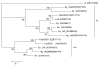
Results: A total of 4111 high quality sequences were obtained from the two libraries and assembled into 370 contigs and 1085 singletons. Molecules with putative roles in blood meal digestion, peritrophic matrix formation, immunity and response to oxidative stress were identified, including proteins that were not previously reported in sand flies. These molecules were evaluated relative to other published sand fly transcripts. Comparative analysis of the two libraries revealed transcripts differentially expressed in response to blood feeding. Molecules up regulated by blood feeding include a putative peritrophin (PperPer1), two chymotrypsin-like proteins (PperChym1 and PperChym2), a putative trypsin (PperTryp3) and four putative microvillar proteins (PperMVP1, 2, 4 and 5). Additionally, several transcripts were more abundant in the sugar fed midgut, such as two putative trypsins (PperTryp1 and PperTryp2), a chymotrypsin (PperChym3) and a microvillar protein (PperMVP3). We performed a detailed temporal expression profile analysis of the putative trypsin transcripts using qPCR and confirmed the expression of blood-induced and blood-repressed trypsins. Trypsin expression was measured in Leishmania infantum-infected and uninfected sand flies, which identified the L. infantum-induced down regulation of PperTryp3 at 24 hours post-blood meal.
Conclusion: This midgut tissue-specific transcriptome provides insight into the molecules expressed in the midgut of P. perniciosus, an important vector of visceral leishmaniasis in the Old World. Through the comparative analysis of the libraries we identified molecules differentially expressed during blood meal digestion. Additionally, this study provides a detailed comparison to transcripts of other sand flies. Moreover, our analysis of putative trypsins demonstrated that L. infantum infection can reduce the transcript abundance of trypsin PperTryp3 in the midgut of P. perniciosus.
Figures







References
-
- Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, Kerhornou A, Ivens A, Fraser A, Rajandream MA, Carver T, Norbertczak H, Chillingworth T, Hance Z, Jagels K, Moule S, Ormond D, Rutter S, Squares R, Whitehead S, Rabbinowitsch E, Arrowsmith C, White B, Thurston S, Bringaud F, Baldauf SL, Faulconbridge A, Jeffares D, Depledge DP, Oyola SO, Hilley JD, Brito LO, Tosi LR, Barrell B, Cruz AK, Mottram JC, Smith DF, Berriman M. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39:839–847. doi: 10.1038/ng2053. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

