The effect of measurement error of phenotypes on genome wide association studies
- PMID: 21569388
- PMCID: PMC3224145
- DOI: 10.1186/1471-2164-12-232
The effect of measurement error of phenotypes on genome wide association studies
Abstract
Background: There is an unspoken assumption that imprecision of measurement of phenotypes will not have large systematic effects on the location of significant associations in a genome wide association study (GWAS). In this report, the effects of two independent measurements of the same trait, subcutaneous fat thickness, were examined in GWAS of 940 individuals.
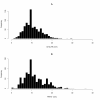
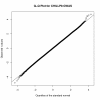
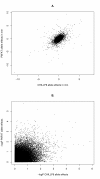
Results: The trait values obtained by two independent groups working to the same trait definition were correlated with r = 0.72. The allele effects obtained from the two analyses were only moderately correlated, with r = 0.53, and there was one significant (P < 0.0001) association in common to the two measurements. The correlation between allele effects was approximately equal to the square of the correlation between the trait measurements. An important quantitative trait locus (QTL) on BTA14 appeared to be shifted distally by 1 Mb along the chromosome. The divergence in GWAS was stronger with data coded into two discrete classes. Univariate trimming of the top and bottom 5% of data, a method used to control for erroneous trait values, decreased the similarity between the GWAS and increased the apparent shift of the QTL on BTA14. Stringent bivariate trimming of data, using only trait values that were similar to each other in the two data sets, substantially improved the correlation of trait values and allele effects in the GWAS, and showed evidence for two QTL on BTA14 separated by 1 Mb. Despite the reduction in sample size due to trimming, more SNP were significant. Using the mean of the two measurements of the trait was not as efficient as bivariate trimming.
Conclusions: It is recommended that trait values in GWAS experiments be examined for repeatability before the experiment is performed. For traits that do not have high repeatability (r < 0.95), two or more independent measurements of the same trait should be obtained for all samples, and individuals genotyped that have highly correlated trait measurements.
Figures


References
-
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ. et al.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

