Comparative proteomics analysis of proteins expressed in the I-1 and I-2 internodes of strawberry stolons
- PMID: 21569547
- PMCID: PMC3113925
- DOI: 10.1186/1477-5956-9-26
Comparative proteomics analysis of proteins expressed in the I-1 and I-2 internodes of strawberry stolons
Abstract
Background: Strawberries (Fragaria ananassa) reproduce asexually through stolons, which have strong tendencies to form adventitious roots at their second node. Understanding how the development of the proximal (I-1) and distal (I-2) internodes of stolons differ should facilitate nursery cultivation of strawberries.
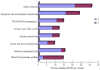
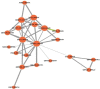
Results: Herein, we compared the proteomic profiles of the strawberry stolon I-1 and I-2 internodes. Proteins extracted from the internodes were separated by two-dimensional gel electrophoresis, and 164 I-1 protein spots and 200 I-2 protein spots were examined further. Using mass spectrometry and database searches, 38 I-1 and 52 I-2 proteins were identified and categorized (8 and 10 groups, respectively) according to their cellular compartmentalization and functionality. Many of the identified proteins are enzymes necessary for carbohydrate metabolism and photosynthesis. Furthermore, identification of proteins that interact revealed that many of the I-2 proteins form a dynamic network during development. Finally, given our results, we present a mechanistic scheme for adventitious root formation of new clonal plants at the second node.
Conclusions: Comparative proteomic analysis of I-1 and I-2 proteins revealed that the ubiquitin-proteasome pathway and sugar-hormone pathways might be important during adventitious root formation at the second node of new clonal plants.
Figures




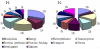


References
-
- Stuefer JF. Separating the effects of assimilate and water integration in clonal fragments by the use of steam-girdling. Abstr Bot. 1995;19:75–81.
-
- Stuefer JF, Huber H. The role of stolon internodes for ramet survival after clone fragmentation in potentilla anserina. Ecol Lett. 1999;2:135–139. doi: 10.1046/j.1461-0248.1999.00066.x. - DOI
-
- Jorrín JD, Rubiales E, Dumas-Gaudot AM, Maldonado M, Castillejo A, Curto M. Proteomics: a promising approach to study biotic interaction in legumes. A review. Euphytica. 2006;147:37–47. doi: 10.1007/s10681-006-3061-1. - DOI
LinkOut - more resources
Full Text Sources

