Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators
- PMID: 21571218
- PMCID: PMC3145975
- DOI: 10.1016/j.devcel.2011.04.008
Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators
Abstract
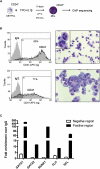
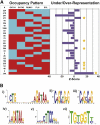
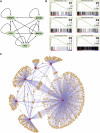
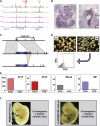
Hematopoietic differentiation critically depends on combinations of transcriptional regulators controlling the development of individual lineages. Here, we report the genome-wide binding sites for the five key hematopoietic transcription factors--GATA1, GATA2, RUNX1, FLI1, and TAL1/SCL--in primary human megakaryocytes. Statistical analysis of the 17,263 regions bound by at least one factor demonstrated that simultaneous binding by all five factors was the most enriched pattern and often occurred near known hematopoietic regulators. Eight genes not previously appreciated to function in hematopoiesis that were bound by all five factors were shown to be essential for thrombocyte and/or erythroid development in zebrafish. Moreover, one of these genes encoding the PDZK1IP1 protein shared transcriptional enhancer elements with the blood stem cell regulator TAL1/SCL. Multifactor ChIP-Seq analysis in primary human cells coupled with a high-throughput in vivo perturbation screen therefore offers a powerful strategy to identify essential regulators of complex mammalian differentiation processes.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


References
-
- Adolfsson J., Mansson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C.T., Bryder D., Yang L., Borge O.J., Thoren L.A. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. - PubMed
-
- Anderson K.P., Crable S.C., Lingrel J.B. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J. Biol. Chem. 1998;273:14347–14354. - PubMed
-
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- G0900729/1/NC3RS_/National Centre for the Replacement, Refinement and Reduction of Animals in Research/United Kingdom
- G0800784/MRC_/Medical Research Council/United Kingdom
- G0900951/MRC_/Medical Research Council/United Kingdom
- 082597/Z/07/Z/WT_/Wellcome Trust/United Kingdom
- RG/09/012/28096/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

