Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs)
- PMID: 21571881
- PMCID: PMC3127695
- DOI: 10.1128/AEM.00468-11
Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs)
Abstract
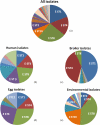
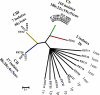
Salmonella enterica subsp. enterica serovar Enteritidis is a major cause of food-borne salmonellosis in the United States. Two major food vehicles for S. Enteritidis are contaminated eggs and chicken meat. Improved subtyping methods are needed to accurately track specific strains of S. Enteritidis related to human salmonellosis throughout the chicken and egg food system. A sequence typing scheme based on virulence genes (fimH and sseL) and clustered regularly interspaced short palindromic repeats (CRISPRs)-CRISPR-including multi-virulence-locus sequence typing (designated CRISPR-MVLST)-was used to characterize 35 human clinical isolates, 46 chicken isolates, 24 egg isolates, and 63 hen house environment isolates of S. Enteritidis. A total of 27 sequence types (STs) were identified among the 167 isolates. CRISPR-MVLST identified three persistent and predominate STs circulating among U.S. human clinical isolates and chicken, egg, and hen house environmental isolates in Pennsylvania, and an ST that was found only in eggs and humans. It also identified a potential environment-specific sequence type. Moreover, cluster analysis based on fimH and sseL identified a number of clusters, of which several were found in more than one outbreak, as well as 11 singletons. Further research is needed to determine if CRISPR-MVLST might help identify the ecological origins of S. Enteritidis strains that contaminate chickens and eggs.
Figures


References
-
- Barrangou R., et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 - PubMed
-
- Beranek A., et al. 2009. Multiple-locus variable-number tandem repeat analysis for subtyping of Salmonella enterica subsp. enterica serovar Enteritidis. Int. J. Med. Microbiol. 299:43–51 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

