Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration
- PMID: 21572092
- PMCID: PMC3108810
- DOI: 10.1093/aob/mcr112
Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration
Abstract
Background and aims: The family of MADS box genes is involved in a number of processes besides controlling floral development. In addition to supplying homeotic functions defined by the ABC model, they influence flowering time and transformation of vegetative meristem into inflorescence meristem, and have functions in roots and leaves. Three Gerbera hybrida At-SOC1-like genes (Gh-SOC1-Gh-SOC3) were identified among gerbera expressed sequence tags.
Methods: Evolutionary relationships between SOC1-like genes from gerbera and other plants were studied by phylogenetic analysis. The function of the gerbera gene Gh-SOC1 in gerbera floral development was studied using expression analysis, protein-protein interaction assays and reverse genetics. Transgenic gerbera lines over-expressing or downregulated for Gh-SOC1 were obtained using Agrobacterium transformation and investigated for their floral phenotype.
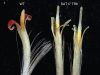
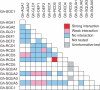
Key results: Phylogenetic analysis revealed that the closest paralogues of At-SOC1 are Gh-SOC2 and Gh-SOC3. Gh-SOC1 is a more distantly related paralogue, grouping together with a number of other At-SOC1 paralogues from arabidopsis and other plant species. Gh-SOC1 is inflorescence abundant and no expression was seen in vegetative parts of the plant. Ectopic expression of Gh-SOC1 did not promote flowering, but disturbed the development of floral organs. The epidermal cells of ray flower petals appeared shorter and their shape was altered. The colour of ray flower petals differed from that of the wild-type petals by being darker red on the adaxial side and greenish on the abaxial surface. Several protein-protein interactions with other gerbera MADS domain proteins were identified.
Conclusions: The At-SOC1 paralogue in gerbera shows a floral abundant expression pattern. A late petal expression might indicate a role in the final stages of flower development. Over-expression of Gh-SOC1 led to partial loss of floral identity, but did not affect flowering time. Lines where Gh-SOC1 was downregulated did not show a phenotype. Several gerbera MADS domain proteins interacted with Gh-SOC1.
Figures





References
-
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, et al. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. The Plant Journal. 2000;24:457–466. - PubMed
-
- Becker A, Theissen G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Molecular Phylogenetics and Evolution. 2003;29:464–489. - PubMed
-
- Borner R, Kampmann G, Chandler J, et al. A MADS domain gene involved in the transition to flowering in Arabidopsis. The Plant Journal. 2000;24:591–599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

