Comparative gene expression analysis throughout the life cycle of Leishmania braziliensis: diversity of expression profiles among clinical isolates
- PMID: 21572980
- PMCID: PMC3091834
- DOI: 10.1371/journal.pntd.0001021
Comparative gene expression analysis throughout the life cycle of Leishmania braziliensis: diversity of expression profiles among clinical isolates
Abstract
Background: Most of the Leishmania genome is reported to be constitutively expressed during the life cycle of the parasite, with a few regulated genes. Inter-species comparative transcriptomics evidenced a low number of species-specific differences related to differentially distributed genes or the differential regulation of conserved genes. It is of uppermost importance to ensure that the observed differences are indeed species-specific and not simply specific of the strains selected for representing the species. The relevance of this concern is illustrated by current study.
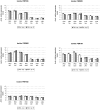
Methodology/principal findings: We selected 5 clinical isolates of L. braziliensis characterized by their diversity of clinical and in vitro phenotypes. Real-time quantitative PCR was performed on promastigote and amastigote life stages to assess gene expression profiles at seven time points covering the whole life cycle. We tested 12 genes encoding proteins with roles in transport, thiol-based redox metabolism, cellular reduction, RNA poly(A)-tail metabolism, cytoskeleton function and ribosomal function. The general trend of expression profiles showed that regulation of gene expression essentially occurs around the stationary phase of promastigotes. However, the genes involved in this phenomenon appeared to vary significantly among the isolates considered.
Conclusion/significance: Our results clearly illustrate the unique character of each isolate in terms of gene expression dynamics. Results obtained on an individual strain are not necessarily representative of a given species. Therefore, extreme care should be taken when comparing the profiles of different species and extrapolating functional differences between them.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Barak E, Amin-Spector S, Gerliak E, Goyard S, Holland N, et al. Differentiation of Leishmania donovani in host-free system: analysis of signal perception and response. Mol Biochem Parasitol. 2005;141:99–108. - PubMed
-
- Haile S, Papadopoulou B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr Opin Microbiol. 2007;10:569–577. - PubMed
-
- Alcolea PJ, Alonso A, Gómez MJ, Moreno I, Domínguez M, et al. Transcriptomics throughout the life cycle of Leishmania infantum: high down-regulation rate in the amastigote stage. Int J Parasitol. 2010;40:1497–1516. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

