Using unsupervised patterns to extract gene regulation relationships for network construction
- PMID: 21573008
- PMCID: PMC3091867
- DOI: 10.1371/journal.pone.0019633
Using unsupervised patterns to extract gene regulation relationships for network construction
Abstract
Background: The gene expression is usually described in the literature as a transcription factor X that regulates the target gene Y. Previously, some studies discovered gene regulations by using information from the biomedical literature and most of them require effort of human annotators to build the training dataset. Moreover, the large amount of textual knowledge recorded in the biomedical literature grows very rapidly, and the creation of manual patterns from literatures becomes more difficult. There is an increasing need to automate the process of establishing patterns.
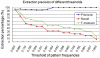
Methodology/principal findings: In this article, we describe an unsupervised pattern generation method called AutoPat. It is a gene expression mining system that can generate unsupervised patterns automatically from a given set of seed patterns. The high scalability and low maintenance cost of the unsupervised patterns could help our system to extract gene expression from PubMed abstracts more precisely and effectively.
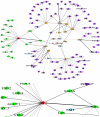
Conclusions/significance: Experiments on several regulators show reasonable precision and recall rates which validate AutoPat's practical applicability. The conducted regulation networks could also be built precisely and effectively. The system in this study is available at http://ikmbio.csie.ncku.edu.tw/AutoPat/.
Conflict of interest statement
Figures






Similar articles
-
A semi-supervised, weighted pattern-learning approach for extraction of gene regulation relationships from scientific literature.Int J Data Min Bioinform. 2014;9(4):401-16. doi: 10.1504/ijdmb.2014.062147. Int J Data Min Bioinform. 2014. PMID: 25757247
-
DMLS: an automated pipeline to extract the Drosophila modular transcription regulators and targets from massive literature articles.Database (Oxford). 2024 Jun 20;2024:0. doi: 10.1093/database/baae049. Database (Oxford). 2024. PMID: 38900628 Free PMC article.
-
Using positive and negative patterns to extract information from journal articles regarding the regulation of a target gene by a transcription factor.Comput Biol Med. 2013 Dec;43(12):2214-21. doi: 10.1016/j.compbiomed.2013.10.011. Epub 2013 Oct 21. Comput Biol Med. 2013. PMID: 24290938
-
Gene association analysis: a survey of frequent pattern mining from gene expression data.Brief Bioinform. 2010 Mar;11(2):210-24. doi: 10.1093/bib/bbp042. Epub 2009 Oct 8. Brief Bioinform. 2010. PMID: 19815645 Review.
-
The impact of post-transcriptional regulation in the p53 network.Brief Funct Genomics. 2013 Jan;12(1):46-57. doi: 10.1093/bfgp/els058. Epub 2012 Dec 14. Brief Funct Genomics. 2013. PMID: 23242178 Free PMC article. Review.
Cited by
-
A Text Mining Protocol for Mining Biological Pathways and Regulatory Networks from Biomedical Literature.Methods Mol Biol. 2022;2496:141-157. doi: 10.1007/978-1-0716-2305-3_8. Methods Mol Biol. 2022. PMID: 35713863
-
AtPAN: an integrated system for reconstructing transcriptional regulatory networks in Arabidopsis thaliana.BMC Genomics. 2012 Mar 8;13:85. doi: 10.1186/1471-2164-13-85. BMC Genomics. 2012. PMID: 22397531 Free PMC article.
References
-
- Blaschke C, Valencia A. The Potential Use of SUISEKI as a Protein Interaction Discovery Tool. Genome Informatics. 2001;12:123–134. - PubMed
-
- Huang M, Zhu X, Hao Y, Payan DG, Qu K, et al. Discovering patterns to extract protein-protein interactions from full texts. Bioinformatics. 2004;20:3604–3612. - PubMed
-
- Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36:664. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

