Crystal structures of two aminoglycoside kinases bound with a eukaryotic protein kinase inhibitor
- PMID: 21573013
- PMCID: PMC3090406
- DOI: 10.1371/journal.pone.0019589
Crystal structures of two aminoglycoside kinases bound with a eukaryotic protein kinase inhibitor
Abstract
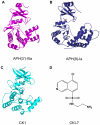
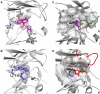
Antibiotic resistance is recognized as a growing healthcare problem. To address this issue, one strategy is to thwart the causal mechanism using an adjuvant in partner with the antibiotic. Aminoglycosides are a class of clinically important antibiotics used for the treatment of serious infections. Their usefulness has been compromised predominantly due to drug inactivation by aminoglycoside-modifying enzymes, such as aminoglycoside phosphotransferases or kinases. These kinases are structurally homologous to eukaryotic Ser/Thr and Tyr protein kinases and it has been shown that some can be inhibited by select protein kinase inhibitors. The aminoglycoside kinase, APH(3')-IIIa, can be inhibited by CKI-7, an ATP-competitive inhibitor for the casein kinase 1. We have determined that CKI-7 is also a moderate inhibitor for the atypical APH(9)-Ia. Here we present the crystal structures of CKI-7-bound APH(3')-IIIa and APH(9)-Ia, the first structures of a eukaryotic protein kinase inhibitor in complex with bacterial kinases. CKI-7 binds to the nucleotide-binding pocket of the enzymes and its binding alters the conformation of the nucleotide-binding loop, the segment homologous to the glycine-rich loop in eukaryotic protein kinases. Comparison of these structures with the CKI-7-bound casein kinase 1 reveals features in the binding pockets that are distinct in the bacterial kinases and could be exploited for the design of a bacterial kinase specific inhibitor. Our results provide evidence that an inhibitor for a subset of APHs can be developed in order to curtail resistance to aminoglycosides.
Conflict of interest statement
Figures
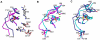
Similar articles
-
Prospects for circumventing aminoglycoside kinase mediated antibiotic resistance.Front Cell Infect Microbiol. 2013 Jun 25;3:22. doi: 10.3389/fcimb.2013.00022. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 23805415 Free PMC article. Review.
-
Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.Biochem J. 2013 Sep 1;454(2):191-200. doi: 10.1042/BJ20130317. Biochem J. 2013. PMID: 23758273 Free PMC article.
-
Inhibition of aminoglycoside antibiotic resistance enzymes by protein kinase inhibitors.J Biol Chem. 1997 Oct 3;272(40):24755-8. doi: 10.1074/jbc.272.40.24755. J Biol Chem. 1997. PMID: 9312069
-
Active site labeling of the gentamicin resistance enzyme AAC(6')-APH(2") by the lipid kinase inhibitor wortmannin.Chem Biol. 2001 Aug;8(8):791-800. doi: 10.1016/s1074-5521(01)00051-5. Chem Biol. 2001. PMID: 11514228
-
Aminoglycoside phosphotransferases: proteins, structure, and mechanism.Front Biosci. 1999 Jan 1;4:D9-21. doi: 10.2741/wright. Front Biosci. 1999. PMID: 9872733 Review.
Cited by
-
Comprehensive review of chemical strategies for the preparation of new aminoglycosides and their biological activities.Chem Soc Rev. 2018 Feb 19;47(4):1189-1249. doi: 10.1039/c7cs00407a. Chem Soc Rev. 2018. PMID: 29296992 Free PMC article. Review.
-
Selective toxicity of antibacterial agents-still a valid concept or do we miss chances and ignore risks?Infection. 2021 Feb;49(1):29-56. doi: 10.1007/s15010-020-01536-y. Epub 2020 Dec 23. Infection. 2021. PMID: 33367978 Free PMC article. Review.
-
Prospects for circumventing aminoglycoside kinase mediated antibiotic resistance.Front Cell Infect Microbiol. 2013 Jun 25;3:22. doi: 10.3389/fcimb.2013.00022. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 23805415 Free PMC article. Review.
-
APH Inhibitors that Reverse Aminoglycoside Resistance in Enterococcus casseliflavus.ChemMedChem. 2025 Apr 14;20(8):e202400842. doi: 10.1002/cmdc.202400842. Epub 2025 Jan 26. ChemMedChem. 2025. PMID: 39801466 Free PMC article.
-
Structural basis for the substrate recognition of aminoglycoside 7''-phosphotransferase-Ia from Streptomyces hygroscopicus.Acta Crystallogr F Struct Biol Commun. 2019 Sep 1;75(Pt 9):599-607. doi: 10.1107/S2053230X19011105. Epub 2019 Aug 28. Acta Crystallogr F Struct Biol Commun. 2019. PMID: 31475927 Free PMC article.
References
-
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2009;48:1–12. - PubMed
-
- De Pascale G, Wright GD. Antibiotic resistance by enzyme inactivation: from mechanisms to solutions. Chembiochem. 2010;11:1325–1334. - PubMed
-
- Therrien C, Levesque RC. Molecular basis of antibiotic resistance and beta-lactamase inhibition by mechanism-based inactivators: perspectives and future directions. FEMS Microbiol Rev. 2000;24:251–262. - PubMed
-
- Burk DL, Berghuis AM. Protein kinase inhibitors and antibiotic resistance. Pharmacol Ther. 2002;93:283–292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

