Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro
- PMID: 21573071
- PMCID: PMC3089636
- DOI: 10.1371/journal.pone.0019711
Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro
Abstract
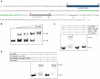
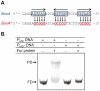
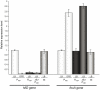
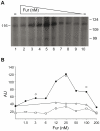
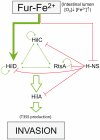
Previous studies have established that the expression of Salmonella enterica pathogenicity island 1 (SPI1), which is essential for epithelial invasion, is mainly regulated by the HilD protein. The ferric uptake regulator, Fur, in turn modulates the expression of the S. enterica hilD gene, albeit through an unknown mechanism. Here we report that S. enterica Fur, in its metal-bound form, specifically binds to an AT-rich region (BoxA), located upstream of the hilD promoter (P(hilD)), at position -191 to -163 relative to the hilD transcription start site. Furthermore, in a P(hilD) variant with mutations in BoxA, P(hilD*), Fur·Mn(2+) binding is impaired. In vivo experiments using S. enterica strains carrying wild-type P(hilD) or the mutant variant P(hilD*) showed that Fur activates hilD expression, while in vitro experiments revealed that the Fur·Mn(2+) protein is sufficient to increase hilD transcription. Together, these results present the first evidence that Fur·Mn(2+), by binding to the upstream BoxA sequence, directly stimulates the expression of hilD in S. enterica.
Conflict of interest statement
Figures


Similar articles
-
Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD.J Bacteriol. 2008 Jan;190(2):476-86. doi: 10.1128/JB.00926-07. Epub 2007 Nov 9. J Bacteriol. 2008. PMID: 17993530 Free PMC article.
-
HilE Regulates HilD by Blocking DNA Binding in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2018 Mar 26;200(8):e00750-17. doi: 10.1128/JB.00750-17. Print 2018 Apr 15. J Bacteriol. 2018. PMID: 29378886 Free PMC article.
-
Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion.J Bacteriol. 2007 Oct;189(19):6882-90. doi: 10.1128/JB.00905-07. Epub 2007 Aug 3. J Bacteriol. 2007. PMID: 17675384 Free PMC article.
-
Regulation of Salmonella enterica pathogenicity island 1 by DNA adenine methylation.Genetics. 2010 Mar;184(3):637-49. doi: 10.1534/genetics.109.108985. Epub 2009 Dec 14. Genetics. 2010. PMID: 20008574 Free PMC article.
-
Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium.Curr Opin Microbiol. 2007 Feb;10(1):24-9. doi: 10.1016/j.mib.2006.12.002. Epub 2007 Jan 5. Curr Opin Microbiol. 2007. PMID: 17208038 Review.
Cited by
-
Interplay between Rho, H-NS, spurious transcription, and Salmonella gene regulation.Proc Natl Acad Sci U S A. 2022 Aug 16;119(33):e2211222119. doi: 10.1073/pnas.2211222119. Epub 2022 Aug 8. Proc Natl Acad Sci U S A. 2022. PMID: 35939681 Free PMC article. No abstract available.
-
PhoP-Mediated Repression of the SPI1 Type 3 Secretion System in Salmonella enterica Serovar Typhimurium.J Bacteriol. 2019 Jul 24;201(16):e00264-19. doi: 10.1128/JB.00264-19. Print 2019 Aug 15. J Bacteriol. 2019. PMID: 31182495 Free PMC article.
-
Functional Insights From KpfR, a New Transcriptional Regulator of Fimbrial Expression That Is Crucial for Klebsiella pneumoniae Pathogenicity.Front Microbiol. 2021 Jan 21;11:601921. doi: 10.3389/fmicb.2020.601921. eCollection 2020. Front Microbiol. 2021. PMID: 33552015 Free PMC article.
-
The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression.PLoS Pathog. 2014 Mar 6;10(3):e1003987. doi: 10.1371/journal.ppat.1003987. eCollection 2014 Mar. PLoS Pathog. 2014. PMID: 24603858 Free PMC article.
-
Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface.PLoS One. 2012;7(1):e29968. doi: 10.1371/journal.pone.0029968. Epub 2012 Jan 17. PLoS One. 2012. PMID: 22272265 Free PMC article.
References
-
- Pang T, Bhutta ZA, Finlay BB, Altwegg M. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 1995;3:253–255. - PubMed
-
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. - PubMed
-
- Jones BD. J Microbiol 43 Spec No; 2005. Salmonella invasion gene regulation: a story of environmental awareness. pp. 110–117. - PubMed
-
- Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 2007;10:24–29. - PubMed
-
- Mastroeni P, Grant A, Restif O, Maskell D. A dynamic view of the spread and intracellular distribution of Salmonella enterica. Nat Rev Microbiol. 2009;7:73–80. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

