Clonal structure of rapid-onset MDV-driven CD4+ lymphomas and responding CD8+ T cells
- PMID: 21573129
- PMCID: PMC3088711
- DOI: 10.1371/journal.ppat.1001337
Clonal structure of rapid-onset MDV-driven CD4+ lymphomas and responding CD8+ T cells
Abstract
Lymphoid oncogenesis is a life threatening complication associated with a number of persistent viral infections (e.g. EBV and HTLV-1 in humans). With many of these infections it is difficult to study their natural history and the dynamics of tumor formation. Marek's Disease Virus (MDV) is a prevalent α-herpesvirus of poultry, inducing CD4+ TCRαβ+ T cell tumors in susceptible hosts. The high penetrance and temporal predictability of tumor induction raises issues related to the clonal structure of these lymphomas. Similarly, the clonality of responding CD8 T cells that infiltrate the tumor sites is unknown. Using TCRβ repertoire analysis tools, we demonstrated that MDV driven CD4+ T cell tumors were dominated by one to three large clones within an oligoclonal framework of smaller clones of CD4+ T cells. Individual birds had multiple tumor sites, some the result of metastasis (i.e. shared dominant clones) and others derived from distinct clones of transformed cells. The smaller oligoclonal CD4+ cells may represent an anti-tumor response, although on one occasion a low frequency clone was transformed and expanded after culture. Metastatic tumor clones were detected in the blood early during infection and dominated the circulating T cell repertoire, leading to MDV associated immune suppression. We also demonstrated that the tumor-infiltrating CD8+ T cell response was dominated by large oligoclonal expansions containing both "public" and "private" CDR3 sequences. The frequency of CD8+ T cell CDR3 sequences suggests initial stimulation during the early phases of infection. Collectively, our results indicate that MDV driven tumors are dominated by a highly restricted number of CD4+ clones. Moreover, the responding CD8+ T cell infiltrate is oligoclonal indicating recognition of a limited number of MDV antigens. These studies improve our understanding of the biology of MDV, an important poultry pathogen and a natural infection model of virus-induced tumor formation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


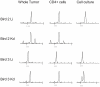

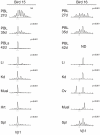




Similar articles
-
Unraveling the role of B cells in the pathogenesis of an oncogenic avian herpesvirus.Proc Natl Acad Sci U S A. 2018 Nov 6;115(45):11603-11607. doi: 10.1073/pnas.1813964115. Epub 2018 Oct 18. Proc Natl Acad Sci U S A. 2018. PMID: 30337483 Free PMC article.
-
Immunomodulation of peripheral T cells in chickens infected with Marek's disease virus: involvement in immunosuppression.J Gen Virol. 1995 Dec;76 ( Pt 12):2979-85. doi: 10.1099/0022-1317-76-12-2979. J Gen Virol. 1995. PMID: 8847503
-
Cytokine gene expression in splenic CD4(+) and CD8(+) T-cell subsets of chickens infected with Marek's disease virus.Viral Immunol. 2009 Feb;22(1):31-8. doi: 10.1089/vim.2008.0062. Viral Immunol. 2009. PMID: 19210226
-
Latency and tumorigenesis in Marek's disease.Avian Dis. 2013 Jun;57(2 Suppl):360-5. doi: 10.1637/10470-121712-Reg.1. Avian Dis. 2013. PMID: 23901747 Review.
-
Revisiting cellular immune response to oncogenic Marek's disease virus: the rising of avian T-cell immunity.Cell Mol Life Sci. 2020 Aug;77(16):3103-3116. doi: 10.1007/s00018-020-03477-z. Epub 2020 Feb 20. Cell Mol Life Sci. 2020. PMID: 32080753 Free PMC article. Review.
Cited by
-
Marek's Disease Virus-Encoded MicroRNA 155 Ortholog Critical for the Induction of Lymphomas Is Not Essential for the Proliferation of Transformed Cell Lines.J Virol. 2019 Aug 13;93(17):e00713-19. doi: 10.1128/JVI.00713-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31189706 Free PMC article.
-
Marek's disease in chickens: a review with focus on immunology.Vet Res. 2016 Nov 28;47(1):119. doi: 10.1186/s13567-016-0404-3. Vet Res. 2016. PMID: 27894330 Free PMC article. Review.
-
Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding.Elife. 2015 Apr 10;4:e05345. doi: 10.7554/eLife.05345. Elife. 2015. PMID: 25860507 Free PMC article.
-
Analysis of the mRNA targetome of microRNAs expressed by Marek's disease virus.mBio. 2014 Jan 21;5(1):e01060-13. doi: 10.1128/mBio.01060-13. mBio. 2014. PMID: 24449754 Free PMC article.
-
Contribution of the TCRβ Repertoire to Marek's Disease Genetic Resistance in the Chicken.Viruses. 2023 Feb 22;15(3):607. doi: 10.3390/v15030607. Viruses. 2023. PMID: 36992316 Free PMC article.
References
-
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. - PubMed
-
- Ishitsuka K, Tamura K. Treatment of adult T-cell leukemia/lymphoma: past, present, and future. Eur J Haematol. 2008;80:185–196. - PubMed
-
- Morrow CaFF. Marek's disease:a worldwide problem. In: Davison TF, Nair V, editors. Marek's disease An evolving Problem. London: Elsevier Academic Press; 2004. pp. 49–61.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

