Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin
- PMID: 21573144
- PMCID: PMC3089418
- DOI: 10.1371/journal.ppat.1001343
Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin
Abstract
The global burden of HIV-associated cryptococcal meningitis is estimated at nearly one million cases per year, causing up to a third of all AIDS-related deaths. Molecular epidemiology constitutes the main methodology for understanding the factors underpinning the emergence of this understudied, yet increasingly important, group of pathogenic fungi. Cryptococcus species are notable in the degree that virulence differs amongst lineages, and highly-virulent emerging lineages are changing patterns of human disease both temporally and spatially. Cryptococcus neoformans variety grubii (Cng, serotype A) constitutes the most ubiquitous cause of cryptococcal meningitis worldwide, however patterns of molecular diversity are understudied across some regions experiencing significant burdens of disease. We compared 183 clinical and environmental isolates of Cng from one such region, Thailand, Southeast Asia, against a global MLST database of 77 Cng isolates. Population genetic analyses showed that Thailand isolates from 11 provinces were highly homogenous, consisting of the same genetic background (globally known as VNI) and exhibiting only ten nearly identical sequence types (STs), with three (STs 44, 45 and 46) dominating our sample. This population contains significantly less diversity when compared against the global population of Cng, specifically Africa. Genetic diversity in Cng was significantly subdivided at the continental level with nearly half (47%) of the global STs unique to a genetically diverse and recombining population in Botswana. These patterns of diversity, when combined with evidence from haplotypic networks and coalescent analyses of global populations, are highly suggestive of an expansion of the Cng VNI clade out of Africa, leading to a limited number of genotypes founding the Asian populations. Divergence time testing estimates the time to the most common ancestor between the African and Asian populations to be 6,920 years ago (95% HPD 122.96 - 27,177.76). Further high-density sampling of global Cng STs is now necessary to resolve the temporal sequence underlying the global emergence of this human pathogen.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

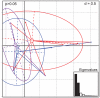

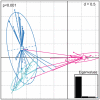


Similar articles
-
Molecular epidemiology of Italian clinical Cryptococcus neoformans var. grubii isolates.Med Mycol. 2013 Jul;51(5):499-506. doi: 10.3109/13693786.2012.751642. Epub 2013 Jan 4. Med Mycol. 2013. PMID: 23286351
-
Molecular epidemiology reveals genetic diversity among 363 isolates of the Cryptococcus neoformans and Cryptococcus gattii species complex in 61 Ivorian HIV-positive patients.Mycoses. 2016 Dec;59(12):811-817. doi: 10.1111/myc.12539. Epub 2016 Jul 27. Mycoses. 2016. PMID: 27461533
-
MLST-Based Population Genetic Analysis in a Global Context Reveals Clonality amongst Cryptococcus neoformans var. grubii VNI Isolates from HIV Patients in Southeastern Brazil.PLoS Negl Trop Dis. 2017 Jan 18;11(1):e0005223. doi: 10.1371/journal.pntd.0005223. eCollection 2017 Jan. PLoS Negl Trop Dis. 2017. PMID: 28099434 Free PMC article.
-
Cryptococcal meningitis due to Cryptococcus neoformans genotype AFLP1/VNI in Iran: a review of the literature.Mycoses. 2015 Dec;58(12):689-93. doi: 10.1111/myc.12415. Epub 2015 Oct 7. Mycoses. 2015. PMID: 26444438 Review.
-
Epidemiology and management of cryptococcal meningitis: developments and challenges.Expert Opin Pharmacother. 2008 Mar;9(4):551-60. doi: 10.1517/14656566.9.4.551. Expert Opin Pharmacother. 2008. PMID: 18312157 Review.
Cited by
-
Population diversity and virulence characteristics of Cryptococcus neoformans/C. gattii species complexes isolated during the pre-HIV-pandemic era.PLoS Negl Trop Dis. 2020 Oct 5;14(10):e0008651. doi: 10.1371/journal.pntd.0008651. eCollection 2020 Oct. PLoS Negl Trop Dis. 2020. PMID: 33017391 Free PMC article.
-
Resistance of Asian Cryptococcus neoformans serotype A is confined to few microsatellite genotypes.PLoS One. 2012;7(3):e32868. doi: 10.1371/journal.pone.0032868. Epub 2012 Mar 13. PLoS One. 2012. PMID: 22427900 Free PMC article.
-
Analyses of the Global Multilocus Genotypes of the Human Pathogenic Yeast Cryptococcus neoformans Species Complex.Genes (Basel). 2022 Nov 6;13(11):2045. doi: 10.3390/genes13112045. Genes (Basel). 2022. PMID: 36360282 Free PMC article.
-
Cryptococcus neoformans Causing Meningoencephalitis in Adults and a Child from Lima, Peru: Genotypic Diversity and Antifungal Susceptibility.J Fungi (Basel). 2022 Dec 16;8(12):1306. doi: 10.3390/jof8121306. J Fungi (Basel). 2022. PMID: 36547639 Free PMC article.
-
Genotypic analysis of clinical and environmental Cryptococcus neoformans isolates from Brazil reveals the presence of VNB isolates and a correlation with biological factors.PLoS One. 2018 Mar 5;13(3):e0193237. doi: 10.1371/journal.pone.0193237. eCollection 2018. PLoS One. 2018. PMID: 29505557 Free PMC article.
References
-
- King J, Dasgupta A. Cryptococcosis. Updated 30th October, 2009. 2005. Available: http://emedicine.medscape.com/article/215354-overview. Accessed 24 April 2010.
-
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. - PubMed
-
- Banerjee U, Datta K, Majumdar T, Gupta K. Cryptococcosis in India: the awakening of a giant? Med Mycol. 2001;39:51–67. - PubMed
-
- Stevens DA, Denning DW, Shatsky S, Armstrong RW, Adler JD, et al. Cryptococcal meningitis in the immunocompromised host: intracranial hypertension and other complications. Mycopathologia. 1999;146:1–8. - PubMed

