Genome-wide mapping of DNA methylation in chicken
- PMID: 21573164
- PMCID: PMC3088676
- DOI: 10.1371/journal.pone.0019428
Genome-wide mapping of DNA methylation in chicken
Abstract
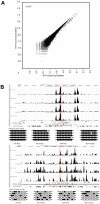
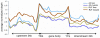
Cytosine DNA methylation is an important epigenetic modification termed as the fifth base that functions in diverse processes. Till now, the genome-wide DNA methylation maps of many organisms has been reported, such as human, Arabidopsis, rice and silkworm, but the methylation pattern of bird remains rarely studied. Here we show the genome-wide DNA methylation map of bird, using the chicken as a model organism and an immunocapturing approach followed by high-throughput sequencing. In both of the red jungle fowl and the avian broiler, DNA methylation was described separately for the liver and muscle tissue. Generally, chicken displays analogous methylation pattern with that of animals and plants. DNA methylation is enriched in the gene body regions and the repetitive sequences, and depleted in the transcription start site (TSS) and the transcription termination site (TTS). Most of the CpG islands in the chicken genome are kept in unmethylated state. Promoter methylation is negatively correlated with the gene expression level, indicating its suppressive role in regulating gene transcription. This work contributes to our understanding of epigenetics in birds.
Conflict of interest statement
Figures

Similar articles
-
Genome-wide DNA methylome variation in two genetically distinct chicken lines using MethylC-seq.BMC Genomics. 2015 Oct 23;16:851. doi: 10.1186/s12864-015-2098-8. BMC Genomics. 2015. PMID: 26497311 Free PMC article.
-
Genome-wide analysis of DNA methylation in bovine placentas.BMC Genomics. 2014 Jan 8;15:12. doi: 10.1186/1471-2164-15-12. BMC Genomics. 2014. PMID: 24397284 Free PMC article.
-
DNA methylation and chromatin structure: the puzzling CpG islands.J Cell Biochem. 2005 Feb 1;94(2):257-65. doi: 10.1002/jcb.20325. J Cell Biochem. 2005. PMID: 15546139 Review.
-
DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter.Hum Mol Genet. 2011 Nov 15;20(22):4299-310. doi: 10.1093/hmg/ddr356. Epub 2011 Aug 11. Hum Mol Genet. 2011. PMID: 21835883 Free PMC article.
-
Functions of DNA methylation: islands, start sites, gene bodies and beyond.Nat Rev Genet. 2012 May 29;13(7):484-92. doi: 10.1038/nrg3230. Nat Rev Genet. 2012. PMID: 22641018 Review.
Cited by
-
Genome-wide analysis of DNA methylation in obese, lean, and miniature pig breeds.Sci Rep. 2016 Jul 22;6:30160. doi: 10.1038/srep30160. Sci Rep. 2016. PMID: 27444743 Free PMC article.
-
Comparison of the genome-wide DNA methylation profiles between fast-growing and slow-growing broilers.PLoS One. 2013;8(2):e56411. doi: 10.1371/journal.pone.0056411. Epub 2013 Feb 18. PLoS One. 2013. PMID: 23441189 Free PMC article.
-
Characteristics of Antisense Transcript Promoters and the Regulation of Their Activity.Int J Mol Sci. 2015 Dec 23;17(1):9. doi: 10.3390/ijms17010009. Int J Mol Sci. 2015. PMID: 26703594 Free PMC article. Review.
-
A chicken DNA methylation clock for the prediction of broiler health.Commun Biol. 2021 Jan 18;4(1):76. doi: 10.1038/s42003-020-01608-7. Commun Biol. 2021. PMID: 33462334 Free PMC article.
-
Analysis of age-related global DNA methylation in chicken.Biochem Genet. 2013 Aug;51(7-8):554-63. doi: 10.1007/s10528-013-9586-9. Epub 2013 Apr 4. Biochem Genet. 2013. PMID: 23553491 Free PMC article.
References
-
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. - PubMed
-
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. - PubMed
-
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

