Identification of hammerhead ribozymes in all domains of life reveals novel structural variations
- PMID: 21573207
- PMCID: PMC3088659
- DOI: 10.1371/journal.pcbi.1002031
Identification of hammerhead ribozymes in all domains of life reveals novel structural variations
Abstract
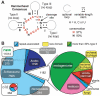
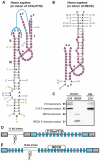
Hammerhead ribozymes are small self-cleaving RNAs that promote strand scission by internal phosphoester transfer. Comparative sequence analysis was used to identify numerous additional representatives of this ribozyme class than were previously known, including the first representatives in fungi and archaea. Moreover, we have uncovered the first natural examples of "type II" hammerheads, and our findings reveal that this permuted form occurs in bacteria as frequently as type I and III architectures. We also identified a commonly occurring pseudoknot that forms a tertiary interaction critical for high-speed ribozyme activity. Genomic contexts of many hammerhead ribozymes indicate that they perform biological functions different from their known role in generating unit-length RNA transcripts of multimeric viroid and satellite virus genomes. In rare instances, nucleotide variation occurs at positions within the catalytic core that are otherwise strictly conserved, suggesting that core mutations are occasionally tolerated or preferred.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs.Genome Biol. 2016 Jun 23;17(1):135. doi: 10.1186/s13059-016-1002-4. Genome Biol. 2016. PMID: 27339130 Free PMC article.
-
Numerous small hammerhead ribozyme variants associated with Penelope-like retrotransposons cleave RNA as dimers.RNA Biol. 2017 Nov 2;14(11):1499-1507. doi: 10.1080/15476286.2016.1251002. Epub 2017 Nov 3. RNA Biol. 2017. PMID: 27858507 Free PMC article.
-
Hammerhead Ribozymes in Archaeal Genomes: A Computational Hunt.Interdiscip Sci. 2017 Jun;9(2):192-204. doi: 10.1007/s12539-016-0141-3. Epub 2016 Jan 13. Interdiscip Sci. 2017. PMID: 26758619
-
Catalytic strategies of self-cleaving ribozymes.Acc Chem Res. 2008 Aug;41(8):1027-35. doi: 10.1021/ar800050c. Epub 2008 Jul 25. Acc Chem Res. 2008. PMID: 18652494 Review.
-
Ribozymes: from mechanistic studies to applications in vivo.J Biochem. 1995 Aug;118(2):251-8. doi: 10.1093/oxfordjournals.jbchem.a124899. J Biochem. 1995. PMID: 8543555 Review.
Cited by
-
An expanded class of histidine-accepting viral tRNA-like structures.RNA. 2021 Jun;27(6):653-664. doi: 10.1261/rna.078550.120. Epub 2021 Apr 2. RNA. 2021. PMID: 33811147 Free PMC article.
-
Identification of Hammerhead-variant ribozyme sequences in SARS-CoV-2.Nucleic Acids Res. 2024 Apr 12;52(6):3262-3277. doi: 10.1093/nar/gkae037. Nucleic Acids Res. 2024. PMID: 38296822 Free PMC article.
-
Discovery of natural non-circular permutations in non-coding RNAs.Nucleic Acids Res. 2023 Apr 11;51(6):2850-2861. doi: 10.1093/nar/gkad137. Nucleic Acids Res. 2023. PMID: 36912096 Free PMC article.
-
A widespread self-cleaving ribozyme class is revealed by bioinformatics.Nat Chem Biol. 2014 Jan;10(1):56-60. doi: 10.1038/nchembio.1386. Epub 2013 Nov 17. Nat Chem Biol. 2014. PMID: 24240507 Free PMC article.
-
Oligonucleotide-Based Approaches to Inhibit Dengue Virus Replication.Molecules. 2021 Feb 11;26(4):956. doi: 10.3390/molecules26040956. Molecules. 2021. PMID: 33670247 Free PMC article. Review.
References
-
- Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–220. - PubMed
-
- Cochrane JC, Strobel SA. Catalytic Strategies of Self-Cleaving Ribozymes. Acc Chem Res. 2008;41:1027–1035. - PubMed
-
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic Processing of Dimeric Plant Virus Satellite RNA. Science. 1986;231:1577–1580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

