No association of xenotropic murine leukemia virus-related viruses with prostate cancer
- PMID: 21573232
- PMCID: PMC3087753
- DOI: 10.1371/journal.pone.0019065
No association of xenotropic murine leukemia virus-related viruses with prostate cancer
Abstract
Background: The association of the xenotropic murine leukemia virus-related virus (XMRV) with prostate cancer continues to receive heightened attention as studies report discrepant XMRV prevalences ranging from zero up to 23%. It is unclear if differences in the diagnostic testing, disease severity, geography, or other factors account for the discordant results. We report here the prevalence of XMRV in a population with well-defined prostate cancers and RNase L polymorphism. We used broadly reactive PCR and Western blot (WB) assays to detect infection with XMRV and related murine leukemia viruses (MLV).
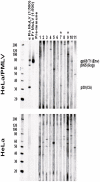
Methodology/principal findings: We studied specimens from 162 US patients diagnosed with prostate cancer with a intermediate to advanced stage (Gleason Scores of 5-10; moderate (46%) poorly differentiated tumors (54%)). Prostate tissue DNA was tested by PCR assays that detect XMRV and MLV variants. To exclude contamination with mouse DNA, we also designed and used a mouse-specific DNA PCR test. Detailed phylogenetic analysis was used to infer evolutionary relationships. RNase L typing showed that 9.3% were homozygous (QQ) for the R462Q RNase L mutation, while 45.6% and 45.1% were homozygous or heterozygous, respectively. Serologic testing was performed by a WB test. Three of 162 (1.9%) prostate tissue DNA were PCR-positive for XMRV and had undetectable mouse DNA. None was homozygous for the QQ mutation. Plasma from all three persons was negative for viral RNA by RT-PCR. All 162 patients were WB negative. Phylogenetic analysis inferred a distinct XMRV.
Conclusions and their significance: We found a very low prevalence of XMRV in prostate cancer patients. Infection was confirmed by phylogenetic analysis and absence of contaminating mouse DNA. The finding of undetectable antibodies and viremia in all three patients may reflect latent infection. Our results do not support an association of XMRV or MLV variants with prostate cancer.
Conflict of interest statement
Figures


References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. - PubMed
-
- Odedina FT, Akinremi TO, Chinegwundoh F, Roberts R, Yu D, et al. Prostate cancer disparities in Black men of African descent: a comparative literature review of prostate cancer burden among Black men in the United States, Caribbean, United Kingdom, and West Africa. Infect Agent Cancer. 2009;4(Suppl 1):S2. - PMC - PubMed
-
- Mo Z, Gao Y, Cao Y, Gao F, Jian L. An updating meta-analysis of the GSTM1, GSTT1, and GSTP1 polymorphisms and prostate cancer: a HuGE review. Prostate. 2009;69:662–688. - PubMed
-
- Murad A, Lewis SJ, Smith GD, Collin SM, Chen L, et al. PTGS2-899G>C and prostate cancer risk: a population-based nested case-control study (ProtecT) and a systematic review with meta-analysis. Prostate Cancer Prostatic Dis 2009 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

